Method for knocking out microRNA gene family by utilizing CRISPR-Cas9 specificity
A gene family, pgem-cas9 technology, applied in the field of specific targeting sgRNA design, can solve the problems of repeated injections, waste of manpower and material resources, restriction of embryos, etc., to achieve simple operation technology, reduced capital investment, and reduced unreliability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] 1. Construct the Cas9 expression vector in vitro, named pGEM-Cas9, the sequence is as shown in SEQ ID NO.1, and the time is 3 days.
[0035] (1) Primer design
[0036] Using the px330 (Addgene:42330) sequence as a template, the primer Cas9-F (the underlined part is the T7 promoter sequence) and the primer Cas9-R were designed.
[0037] Cas9-F:
[0038] 5'- ATGGACTATAAGGACCACGAC-3';
[0039] Cas9-R: 5'-GCGAGCTCTAGGAATTCTTAC-3'.
[0040] (2) PCR amplification
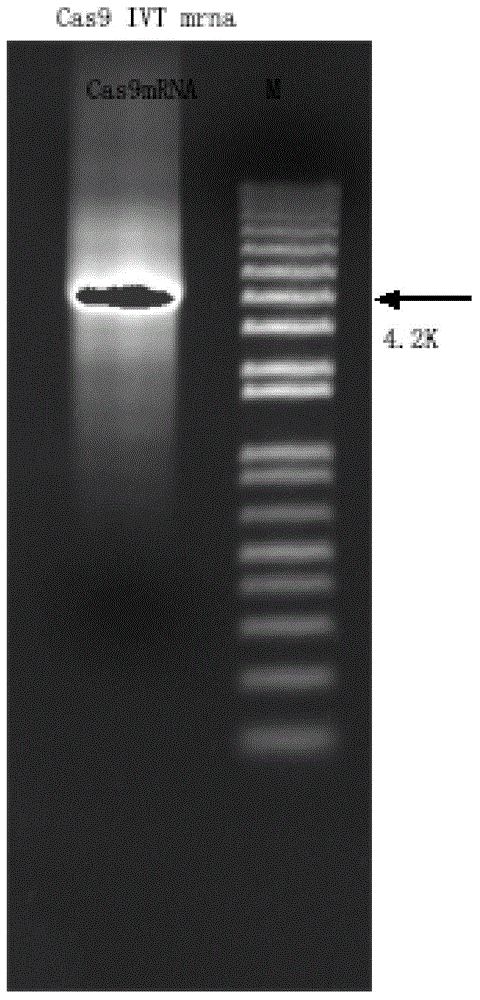
[0041]Using px330 as a template, PCR reaction was carried out under the action of primer Cas9-F and primer Cas9-R, and the amplified product was analyzed by gel electrophoresis, and the target fragment size was 5309bp, such as figure 2 shown.
[0042] The PCR reaction system is:
[0043]
[0044] The PCR reaction conditions are:
[0045]
[0046] (3) Construction of pGEM-Cas9 expression vector
[0047] Take 1 μl of the PCR product of step (2) and connect it to the pGEM-T vector, and place it at roo...
Embodiment 2
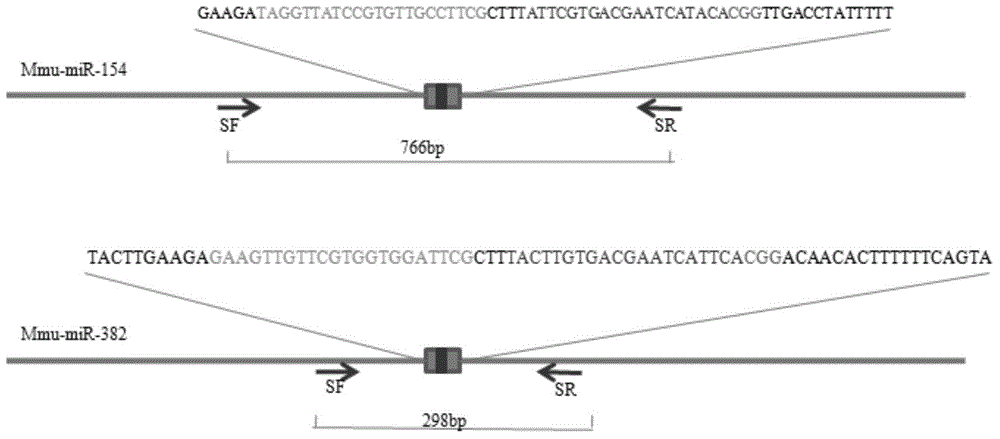
[0076] 1. Design sgRNA for mmu-miR-154 and mmu-miR-382 genes, add a BbsI restriction site (underlined) at the 5' end of the primer, and obtain miR-154 sgRNA (sequence such as SEQ ID NO.3) through denaturation and annealing reactions ) and miR-382sgRNA (sequence such as SEQ ID NO.4), the time is 2 days.
[0077] sgRNA-miR-382-F:5'- TACTTGTGACGAATCATTCA-3'
[0078] sgRNA-miR-382-R:5'- TGAATGATTCGTCACAAGTAC-3'
[0079] sgRNA-miR-154-F:5'- TATTCGTGACGAATCATACA-3'
[0080] sgRNA-miR154-R:5'- TGTATGATTCGTCACGAATAC-3'
[0081] Denaturation, annealing reaction system is as follows:
[0082]
[0083]
[0084] The reaction system in the PCR instrument is as follows: 37°C for 30min; 95°C for 5min; 95-25°C, 5°C / min, and the product is miR-382sgRNA.
[0085] Denaturation, annealing reaction system is as follows:
[0086]
[0087] The reaction system in the PCR instrument was as follows: 37°C, 30min; 95°C, 5min; 95-25°C, 5°C / min, and the product was miR-154sgRNA.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com