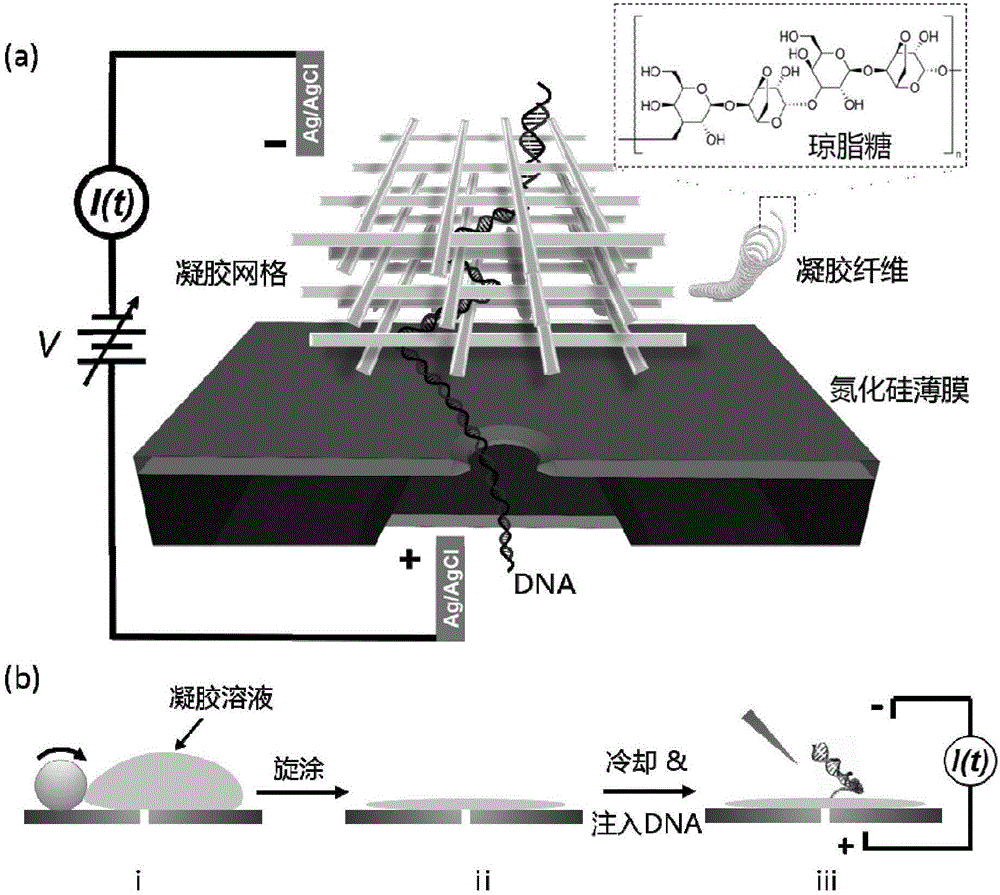
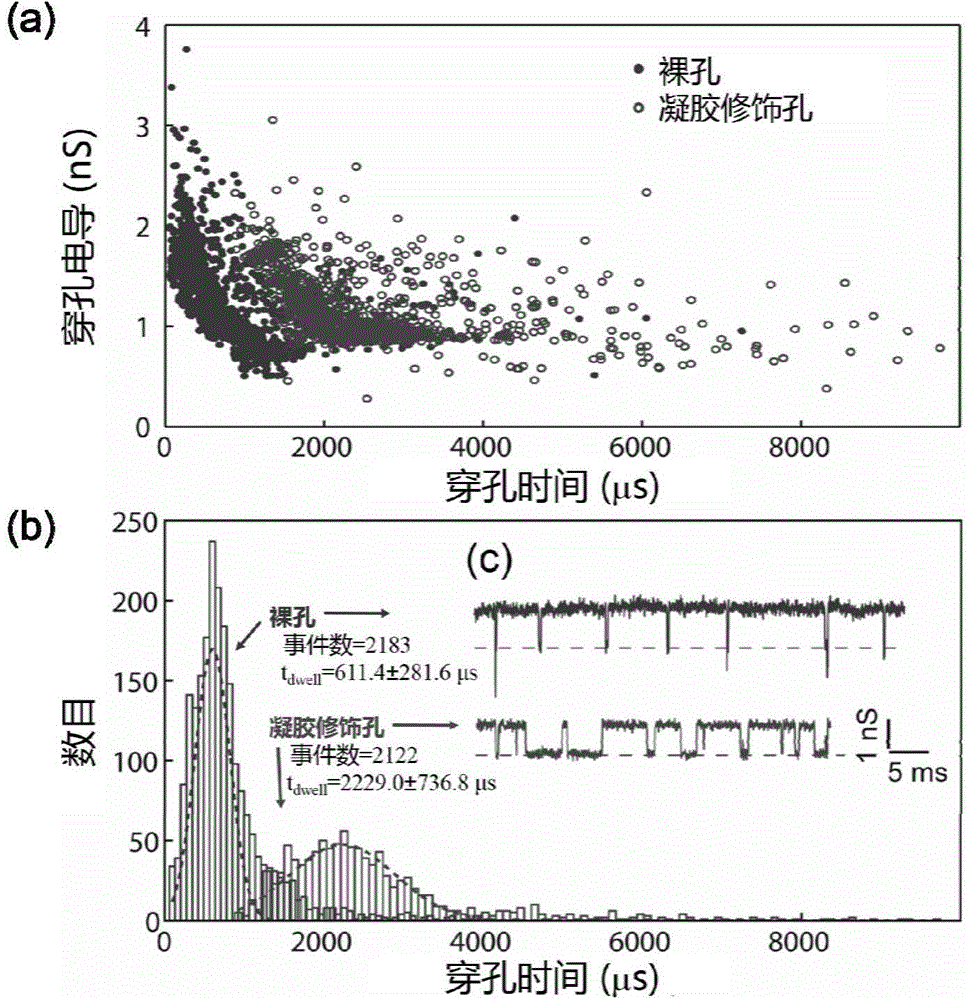
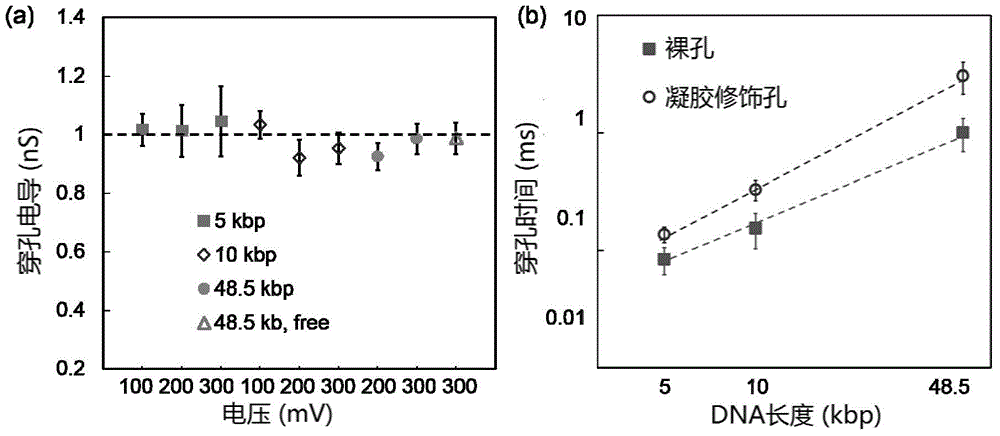
Nanopore sequencing method for nucleic acid molecules with low perforation rate and special nanopore device thereof
A technology of nanopore sequencing and low perforation speed, applied in biochemical equipment and methods, determination/inspection of microorganisms, bioreactors/fermenters for specific purposes, etc. DNA perforation blocks the current value amplitude decrease, and the difficulty of accurately measuring the signal increases, so as to achieve the effect of simple and easy experiment, significant deceleration effect, and improved time resolution.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Method for reducing the perforation speed of nucleic acid molecules in the nanopore sequencing method
[0037] 1. Preparation of chip devices
[0038] A high-energy convergent electron beam in a transmission electron microscope is used to drill holes in the suspended film of the device to prepare a chip device. This includes a series of micro-nano processing technology involved in traditional semiconductor processing technology, and also involves a series of modern cutting-edge nano-scale processing technology.
[0039] The specific method is as follows: First, a 4-inch silicon wafer with a (100) surface is grown with 2 microns of silicon oxide on both sides, and low-stress silicon nitride of about 150 to 200 nanometers is deposited by a low-pressure chemical vapor deposition method. Then make a photolithographic mask, the purpose is to make a lot of 3mm×3mm small substrate periodic distribution on the 4-inch silicon wafer, and each 3mm×3mm small substrate p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com