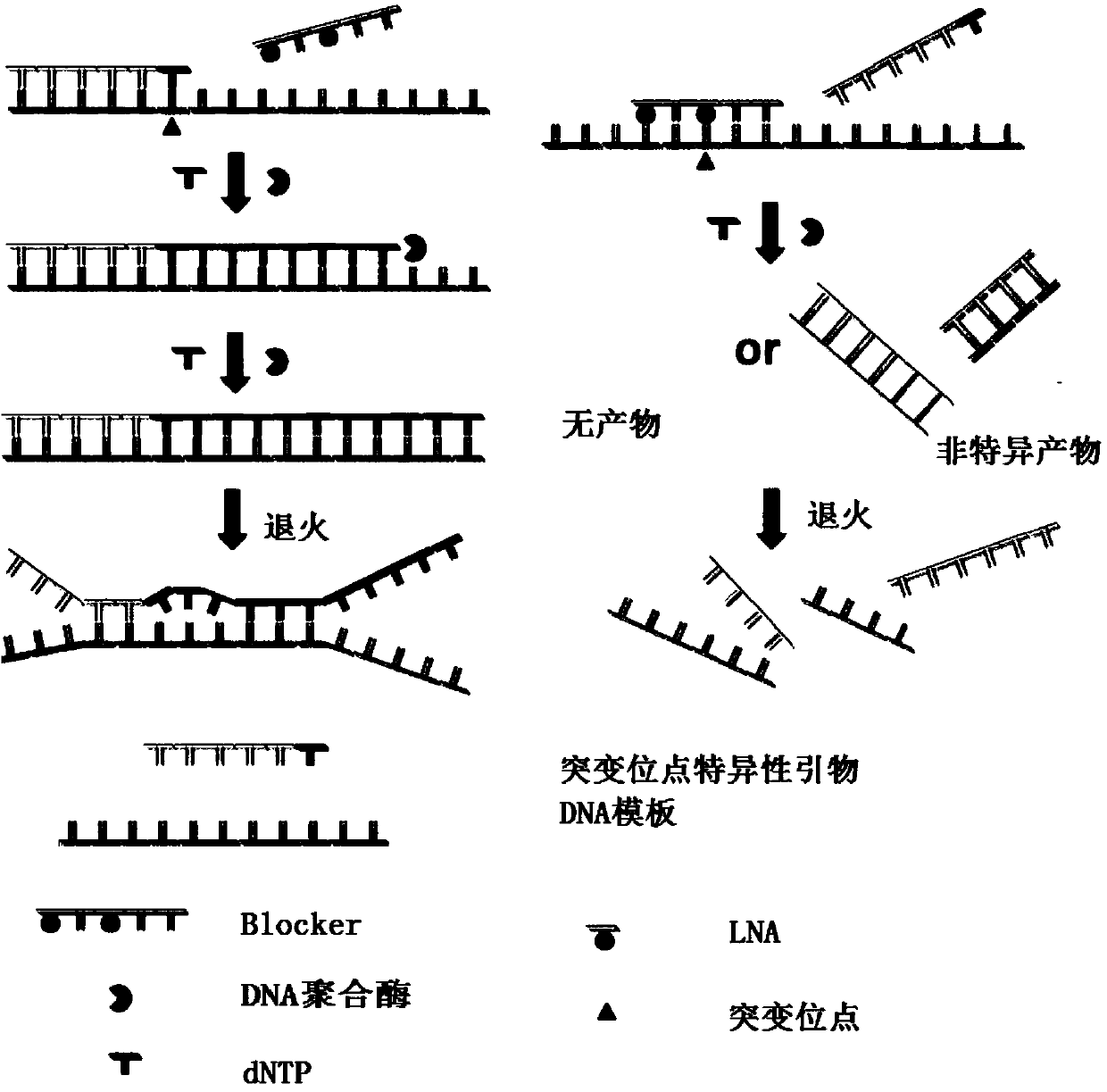
Kit for detecting EGFR gene mutation and detection method thereof
A kit and base technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of easy contamination, positive results can not confirm the specific unknown, etc., to achieve strong specificity, test personnel and environment. No harm, good safety effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The EGFR gene negative quality control product is human serum genomic DNA without EGFR gene mutation, and then diluted to 10 3 copy number. The EGFR gene positive quality control product is connected to the pUC57-Amp vector by the mutation sequence of 29 mutation sites corresponding to the 4 exons of the EGFR gene, EGFR18, EGFR19, EGFR20 and EGFR21. After transformation into Escherichia coli, it is extracted and purified, and finally diluted into Concentration is 10 3 Copy number / μl. The mutation site information comes from the Cosmic database, which are G719A, G719S, G719C, E746_A750del(1), E746_A750del(2), L747_P753>S, E746_T751>I, E746_T751del, E746_T751>A, E746_S752>A, E746_S752 E746_S752>D、L747_A750>P、L747_T751>Q、L747_E749del、L747_T751del、L747_S752del、L747_A750>P、L747_P753>Q、L747_T751>S、L747_T751del、L747_T751>P、T790M、S768I、H773_V774insH、D770_N771insG、V769_D770insASV、L858R、L861Q, Among them, the deletion mutation site on EGFR19 was named 19-del; the insertion mut...
Embodiment 2
[0080] Accurately dilute the 29 mutation-positive quality control products and Control quality control products, and uniformly dilute to 10 3 Copy number, the dilution solution is 10ng / μl wild-type DNA, and then the positive quality control product and the Control quality control product are respectively 0:100, 0.01:99.99, 0.5:99.5 according to the ratio of the positive quality control product and the Control quality control product , 5:95 to prepare sensitivity reference products. Considering that there may be differences caused by adding samples or personal factors during the experiment, we choose to repeat the measurement 3 times, and compare and analyze the mixed sample with 0% each time to test its sensitivity, and analyze the corresponding sensitivity according to the average value in the results interval. According to the results of the minimum detection limit experiment, no amplification occurred in the blank samples of all sites, and the experiment is credible. When...
Embodiment 3
[0086] Collect 100 lung cancer patient tissue samples and matched preoperative plasma samples. The samples are all collected from the First Affiliated Hospital of Zhejiang University School of Medicine. Immediately after sample collection, store in a -80°C refrigerator. The DNA in the tissue samples and the free DNA (cf-DNA) in the plasma were extracted using Qiagen kits, and subjected to agarose gel electrophoresis and concentration determination.
[0087] Then, the method of Example 1 was used to detect mutation sites in the tissue samples and plasma samples of 15 lung cancer patients, and the tissue samples were sequenced at the same time, and the type of mutation was determined according to the sequencing results. The results are shown in Table 9.
[0088] Table 9, adopt the method of the present invention to detect lung cancer patient result
[0089]
[0090] The results show that the coincidence rate of mutations in the plasma samples detected by the method of the pr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com