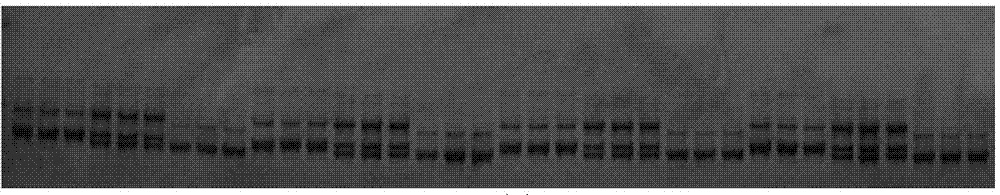
Method for high flux rapid extraction of vegetable single seed DNA
An extraction method and high-throughput technology, applied in the field of molecular biology, can solve the problems of time-consuming, many extraction steps, and complexity, and achieve the effect of saving time, simple process and fast process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Taking the cauliflower variety "Jinpin 70" hybrid as a sample, the method of the present invention was used to rapidly extract DNA. The preparation method is as follows:
[0066] (1) Take about 100 mg of a single cauliflower hybrid seedling and add it to a 1.5 mL sterilized centrifuge tube, and use a hand-held electric crusher to grind it to powder;
[0067] (2) Add 100 μL of 10% Chelex-100 solution, vortex and mix thoroughly, and place in a 65°C water bath for 20 minutes at a constant temperature;
[0068] (3) After the water bath, quickly freeze on ice for 10 minutes, centrifuge at 13,200 rpm / min for 3 minutes, take the supernatant, and save it for subsequent PCR amplification.
[0069] PCR detection:
[0070] 1. Amplify the cauliflower EST-SSR purity identification primer HYC33, the primer sequence is:
[0071] HYC33-F: 5'-AGACTTCGTCGATCCACCTG-3'
[0072] HYC33-R: 5'-GACCTCGTCTCCTCTTCCT-3'
[0073] 2. Reaction system:
[0074] 2× Master Mixes 5 μL, HYC33 upst...
Embodiment 2
[0082] Using the Chinese cabbage variety "Qiulu 75" hybrid as a sample, the method of the present invention was used to rapidly extract DNA. The preparation method is as follows:
[0083] (1) Take about 100 mg of a single Chinese cabbage hybrid seedling and add it to a 1.5 mL sterilized centrifuge tube, and use a hand-held electric crusher to grind it to powder;
[0084] (2) Add 100 μL of 10% Chelex-100 solution, vortex and mix thoroughly, and place in a 65°C water bath for 20 minutes at a constant temperature;
[0085] (3) After the water bath, quickly freeze on ice for 10 minutes, centrifuge at 13,200 rpm / min for 3 minutes, take the supernatant, and save it for subsequent PCR amplification.
[0086] PCR detection:
[0087] 1. Amplify the Chinese cabbage EST-SSR purity identification primer BC1, the primer sequence is:
[0088] BC1-F: 5'-TTTGTCCCTATTGCTCAGGG-3'
[0089] BC1-R: 5'-CCGAGAACGTCTTTCTCTTG-3'
[0090] 2. Reaction system:
[0091] 2× Master Mixes 5 μL, upstr...
Embodiment 3
[0099] Using the Chinese cabbage variety "Jinbai 56" hybrid as a sample, the method of the present invention was used to rapidly extract DNA. The preparation method is as follows:
[0100] (1) Take about 100 mg of a single Chinese cabbage hybrid seedling and add it to a 1.5 mL sterilized centrifuge tube, and use a hand-held electric crusher to grind it to powder;
[0101] (2) Add 100 μL of 10% Chelex-100 solution, vortex and mix thoroughly, and place in a 65°C water bath for 20 minutes at a constant temperature;
[0102] (3) After the water bath, quickly freeze on ice for 10 minutes, centrifuge at 13,200 rpm / min for 3 minutes, take the supernatant, and save it for subsequent PCR amplification.
[0103] PCR detection:
[0104] 1. Amplify the Chinese cabbage EST-SSR purity identification primer BC9, the primer sequence is:
[0105] BC9-F: 5'-ACCTTCCGTCTCTGGGTCTT-3'
[0106] BC9-R: 5'-TTTTGAACGAGGTGGAGGAC-3'
[0107] 2. Reaction system:
[0108] 2× Master Mixes 5 μL, upst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com