Multi-sample and multi-segment DNA methylation high-throughput sequencing method
A methylation and multi-fragment technology, applied in recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc., can solve PCR recombination between alleles, cannot detect methylation status, and consumes a lot of time and money and other issues, to achieve the effect of using sequencing throughput, low incidence of PCR recombination, and sufficient sequencing throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
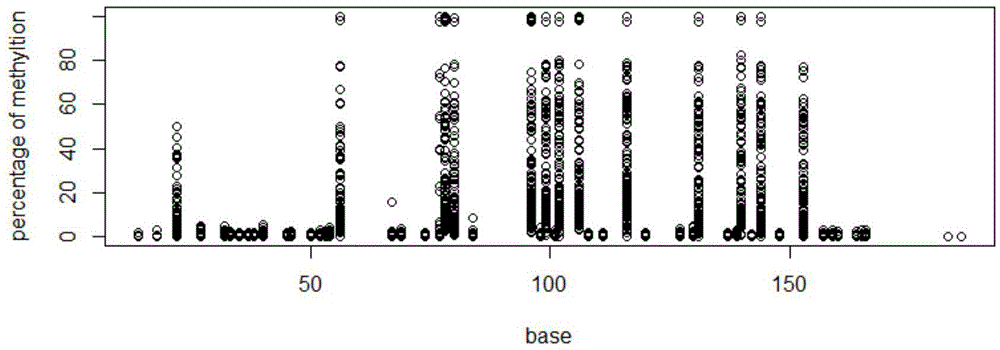
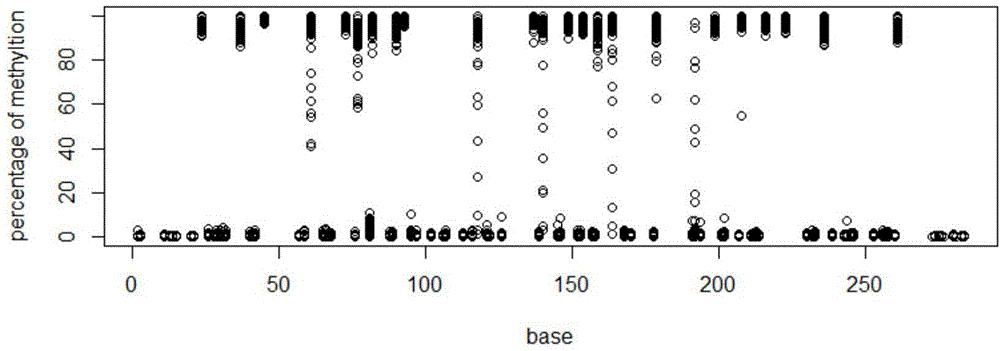
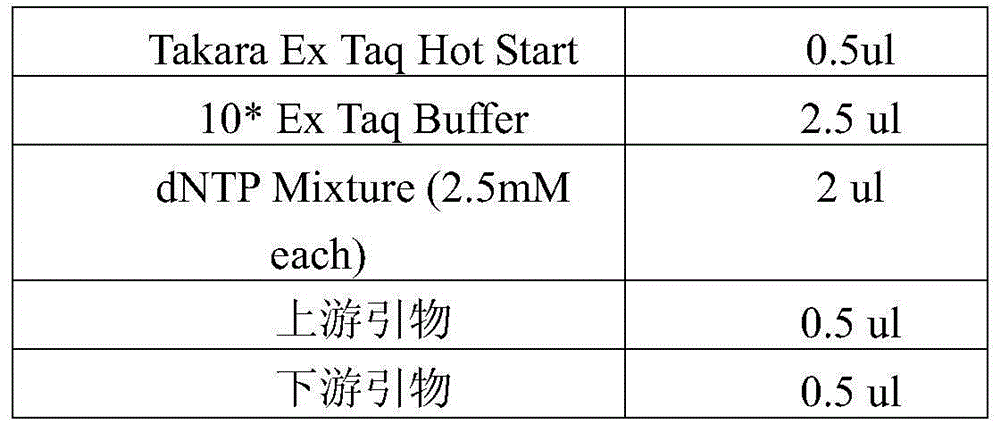
[0029] The main technical process of the present invention: extract genomic DNA, then perform bisulfite treatment on genomic DNA, and then perform PCR amplification on the processed genomic DNA, wherein different PCR primers are used for different gene fragments, and the same gene fragments use The label (barcode) sequence distinguishes different samples, and then mixes different PCR products. The mixed PCR products are prepared for sequencing library, and then the library is subjected to QPCR quality inspection and quantification, then sequenced on the machine, and finally different sample specificity High-precision analysis of fragment methylation status.
[0030] According to an embodiment of the present invention, the sample genomic DNA can be derived from any species, including but not limited to genomic DNA of animals, plants, and insects.
[0031] According to an embodiment of the present invention, the extracted genomic DNA is treated with bisulfite, such as using EZ D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com