Special primer capable of quickly detecting human coronavirus NL63 as well as kit and application method thereof
A coronavirus, human detection technology, applied in the fields of molecular biology and virology, to achieve specific results in the detection process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 special primer design
[0031] The 32 currently known full-length genome sequences of HCoV-NL63 registered on GenBank were analyzed by Clustal software, and the domestic strain (GenBank accession number JX104161) was used as the reference sequence, and PrimerExplorer4.0 software was used to target HCoV-NL63. Eight primers were designed for the coding region of NL63 nucleocapsid (N) gene, namely F3, F2, F1, B1, B2, B3, LF, and LB. Among them, F3 and B3 are two outer primers, FIP (composed of F1 and F2) and BIP (composed of B1 and B2) are two inner primers, and LF and BF are two loop primers. See the table below for primer information:
[0032] F3: CATTTTACATGCCTCTTTTGG
[0033] B3: CCTTATGAGGTCCAGTACC
[0034] FIP: TCCAATAACCAATCTGCTCATCTTTTGATAAGGCACCATATAGGGT
[0035] BIP: GTTCAAGAGCGTTGGCGTATAAAATGAACTTTAGGAGGCAAAT
[0036] LF: GGGACAAGATTCCTGGGAATG
[0037] BF: CAGGGGGCAACGTGTTG.
Embodiment 2
[0038]Example 2 Primer Specificity Test
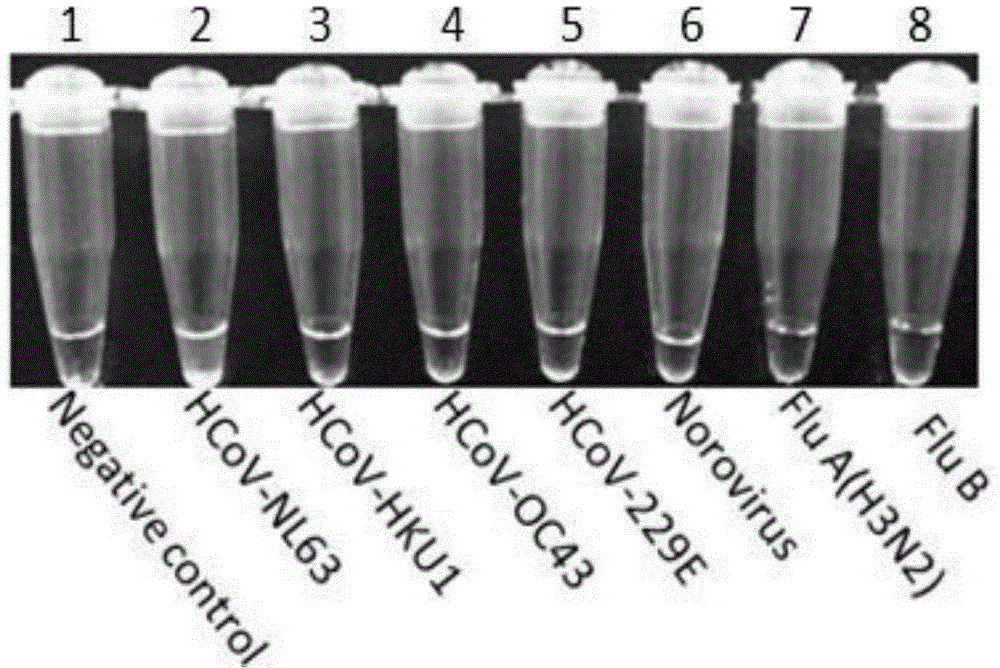
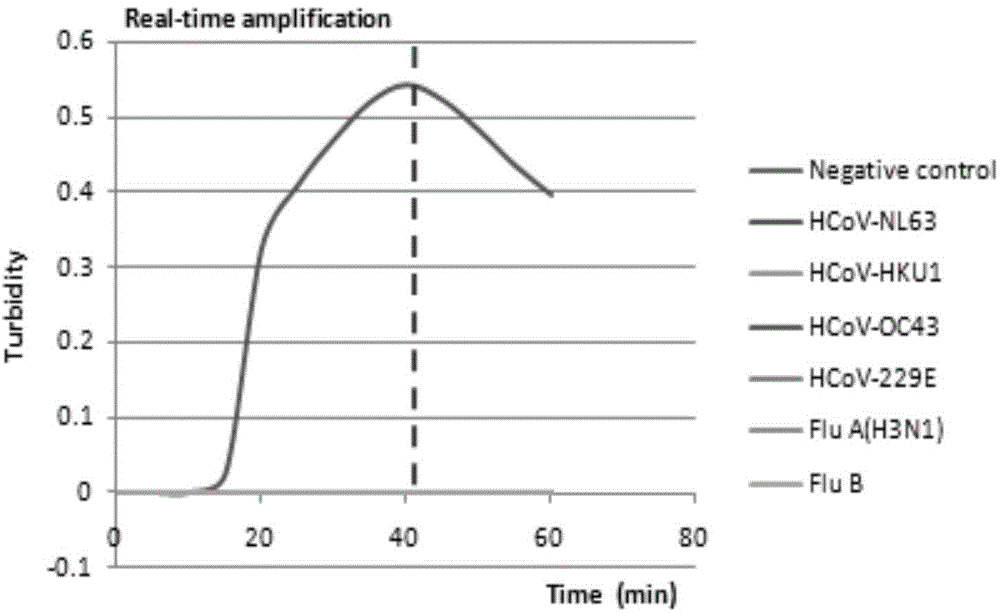
[0039] Four common human coronaviruses, HCoV-NL63, HCoV-OC43, HCoV-229E and HCoV-HKU, as well as influenza A virus (H3N2) and influenza B virus (Flu B), Norovirus (Norovirus) genome was used as a template respectively, and deionized water was used as a negative control, and the following 25 μL reaction system was established according to the RT-LAMP kit: 12.5 μL of 2x reaction buffer (RM), six specific primers ( F3, B3, FIP, BIP, LF, BF) 1 μL each (primer concentrations are 5 μM, 5 μM, 40 μM, 40 μM, 20 μM, 20 μM), visual fluorescence reagent (calcein) 1 μL, enzyme solution (EM) 1 μL, virus 4.5 μL of RNA template was mixed evenly and placed in an LA-500 Loopamp instrument for constant temperature reaction at 63°C for 60 minutes, then at 80°C for 5 minutes to terminate the reaction and observe the color change in the reaction tube. The result is shown in the figure below:
[0040] from figure 1 It can be seen that only in the reaction...
Embodiment 3
[0041] Example 3 Primer Sensitivity Test
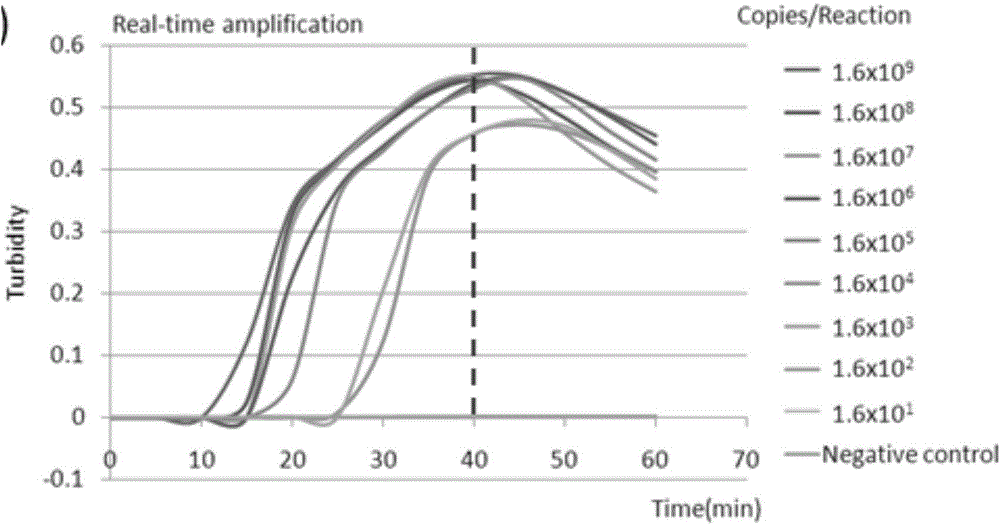
[0042] Use a spectrophotometer to quantify HCoV-NL63 viral RNA, and estimate each microliter according to the formula (copy / μL=(6.02x1023)xconcentration (ng / μL)x10-9 / (target fragment base number x340)) Viral RNA copy number. The RNA was serially diluted to 10 by 10-fold dilution 0 times, 10 1 times, 10 2 times, 10 3 times, 10 4 times, 10 5 times, 10 6 times, 10 7 times, 10 8 times. Establish the following 25 μL RT-LAMP reaction system: 12.5 μL of 2x reaction buffer (RM), 1 μL of each of the six specific primers (F3, B3, FIP, BIP, LF, BF) (primer concentrations were 5 μM, 5 μM, 40 μM, 40 μM, 20 μM, 20 μM), 1 μL of visual fluorescence reagent (calcein), 1 μL of enzyme solution (EM), 5 μL of viral RNA template diluted in each ratio, mix well and place in LA-500 Loopamp instrument for constant temperature reaction at 63°C for 60 min, and Observe the change of reaction turbidity value in real time. After the end of the reaction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com