SNP molecular marker associated with pig fat deposition, and applications thereof
A molecular marker, pig fat technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the lack of major genes and molecular markers, substantial progress, actual identification of major genes, Regulatory pathways and their applications are complex
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] The ear tissue samples of the Chinese local breed "Diannan small-eared pig" (collected from the Diannan small-eared pig resource protection farm in Xishuangbanna, Yunnan Province) and the introduced breed "Darkie pig" (collected from Anhui Kexin Pig Breeding Co., Ltd.) were selected. Genomic DNA was extracted by the phenol / chloroform method.
[0036] Design the following primers (as shown in the sequence listing SEQIDNO.2 and SEQIDNO.3):
[0037] Forward primer F: 5'-ACCTGACCTCCAGCTCTCAAAG-3'
[0038] Reverse primer R: 5'-GCAGTGATGGGGATGATGG-3'
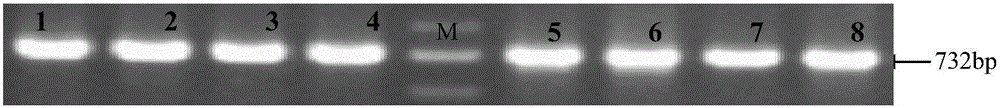
[0039] Use the above primers to carry out PCR amplification on the genomic DNA of small-eared pigs and large grammes in southern Yunnan. PCR amplification uses a 20 μl reaction system: the amount of 2×PCRMix is half of the volume of the PCR amplification reaction system, that is, 10 μl, and 100 ng of the pig genomic DNA to be detected , 10μmol / L forward and reverse primers 0.5μl each, and finally use ddH 2 Add O to 20 μl a...
Embodiment 2
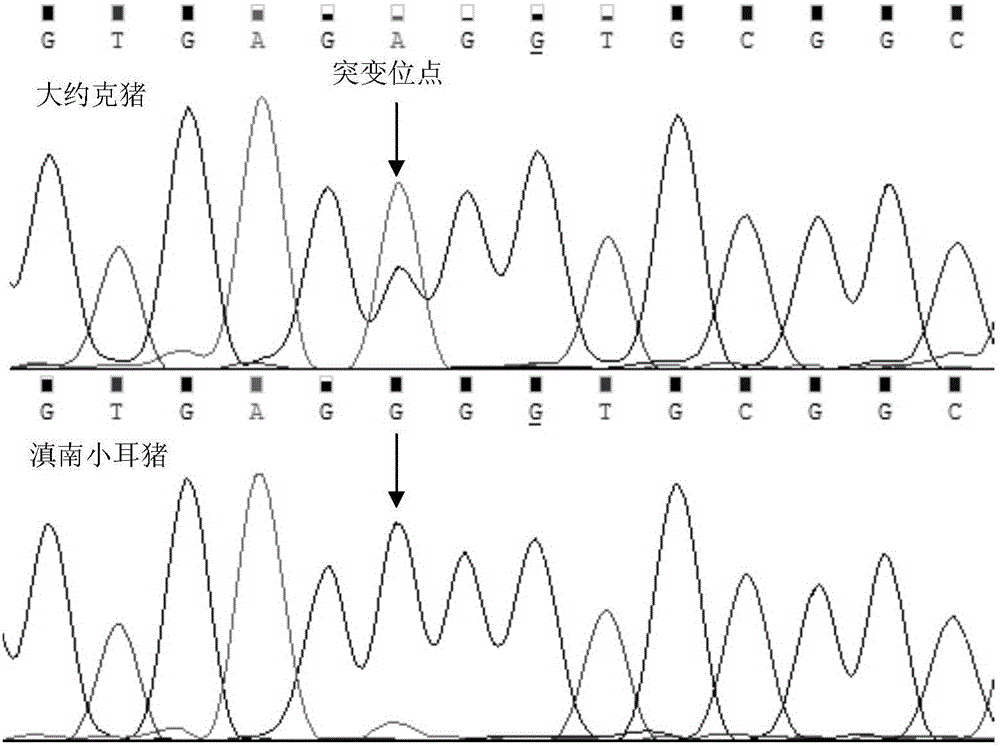
[0043] Establishment of a sequencing method for genotype detection at A-931G site of porcine FNDC5 gene.
[0044] In order to detect the genotype of the A-931G locus of the pig FNDC5 gene quickly and conveniently, the genomic DNA of the pig is subjected to PCR amplification with the primer sequence in Example 1, and the PCR reaction system is a 20 μl system: 2×PCRMix dosage is the PCR amplification reaction Half of the system volume is 10 μl, 100ng of the porcine genomic DNA to be detected, 10μmol / L forward and reverse primers 0.5μl each, and finally use ddH 2Add O to 20 μl and mix well to obtain the PCR amplification reaction system. The PCR amplification reaction system was pre-denatured at 94-95°C for 5 minutes; 36 cycles were performed as follows: pre-denaturation at 94-95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 40 s, and extension at 72°C for 7 min to obtain the PCR product.
[0045] The PCR amplification product was electrophoresed on agarose gel w...
Embodiment 3
[0047] The genotype distribution detection of the SNP site of the present invention in different pig groups.
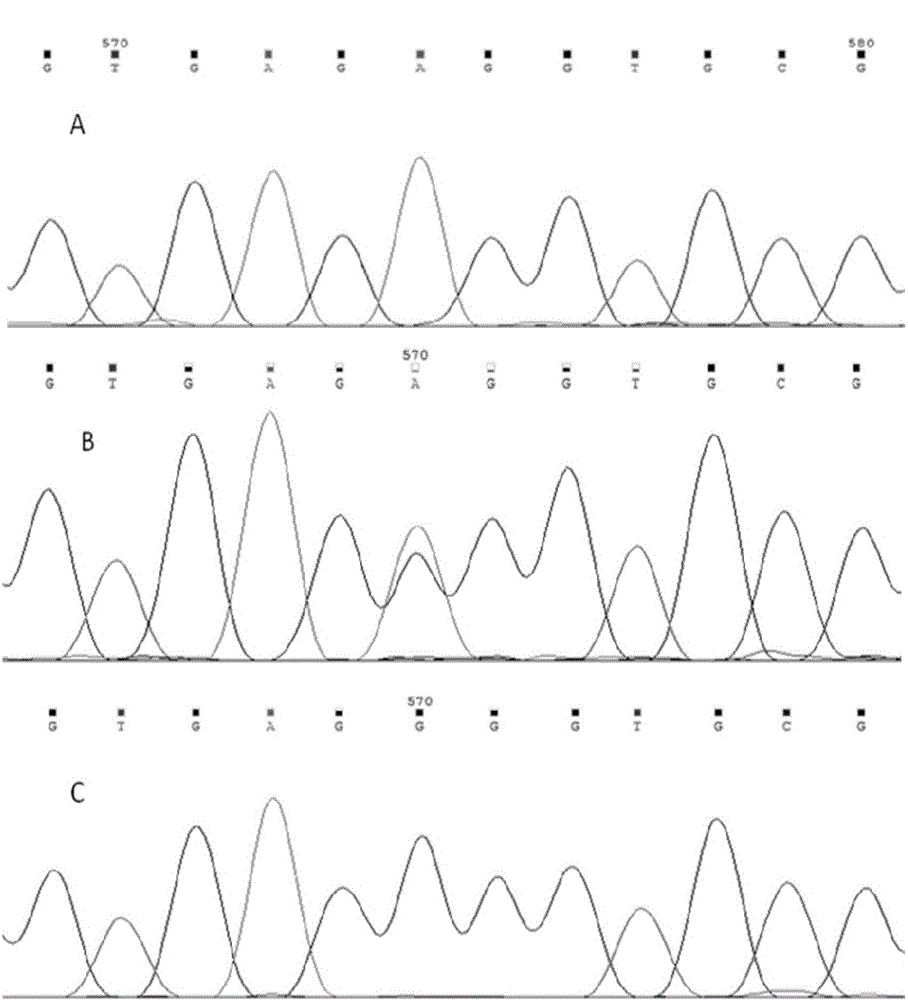
[0048] The ear tissue samples of Tibetan pigs (Bujiangda County, Nyingchi, Tibet), Diannan small-eared pigs (Xishuangbanna, Yunnan), Yorkshire pigs (Anhui Kexin Pig Breeding Co., Ltd.) and Landrace pigs (Beijing Zhongshun Jingsheng Breeding Co., Ltd.) were collected and tested with phenol / chloroform method to extract pig individual genomic DNA samples.
[0049] The genotypes of the A-931G loci of the FNDC5 gene of the individuals of the four varieties were determined by amplified PCR product sequencing technology, and the detection results are shown in Table 1.
[0050] Table 1 Genotype distribution of pig FNDC5 gene A-931G locus
[0051]
[0052] Both Tibetan pigs and Diannan small-eared pigs belong to Chinese local pig breeds with slow growth rate, and their fat deposition ability is stronger than that of imported breed pigs, which belong to fat pigs. It can b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com