Quinoa est-ssr molecular marker and its application
A molecular marker, quinoa technology, used in DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of difficulty in large-scale development of molecular markers and difficulty in genome sequence assembly, and achieve rich polymorphism, good versatility, high success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
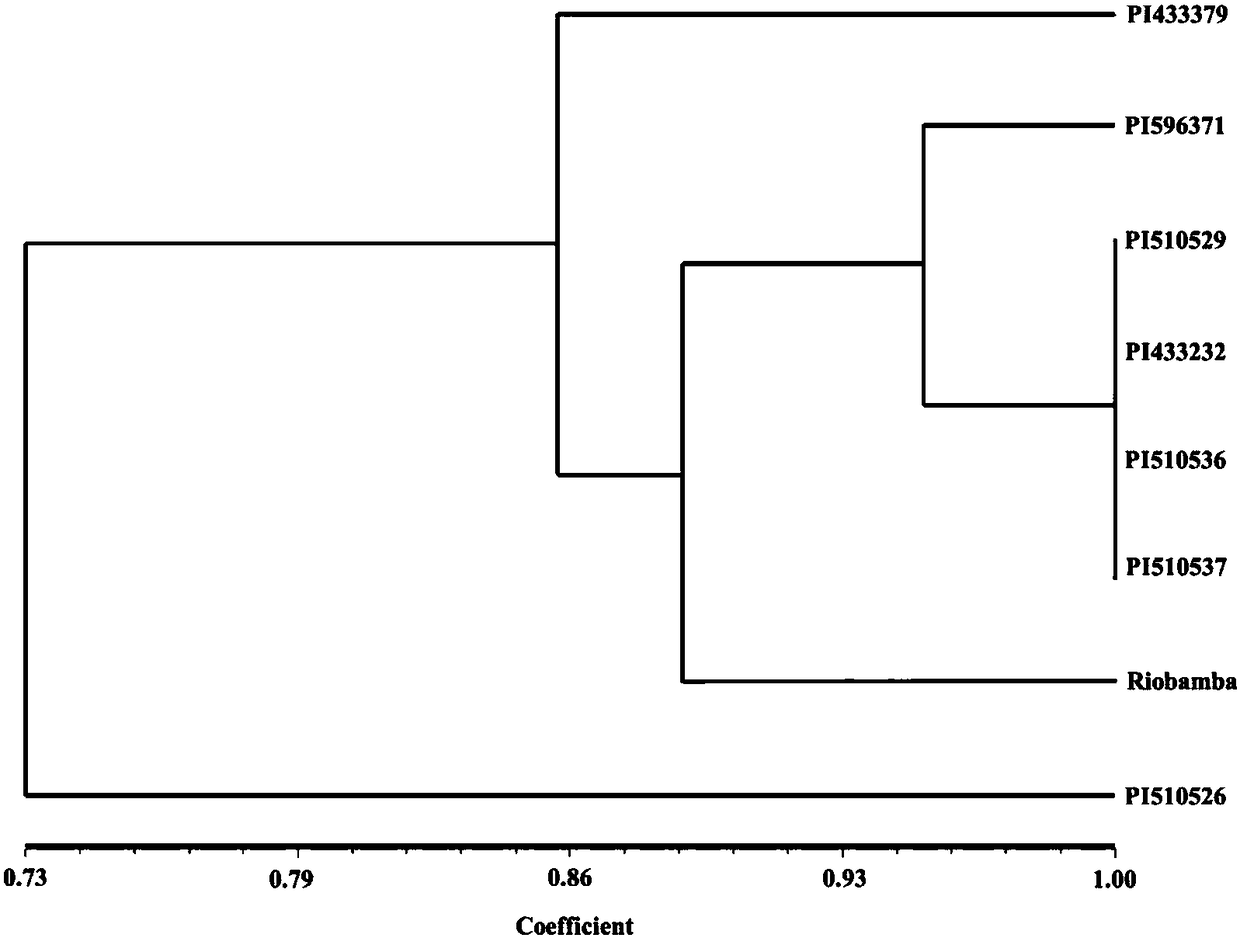
Image
Examples
Embodiment Construction
[0016] The following examples facilitate a better understanding of the present invention, but do not limit the present invention. The experimental methods in the following examples are conventional methods unless otherwise specified. The test materials used in the following examples, unless otherwise specified, were purchased from conventional biochemical reagent stores.
[0017] 1) Acquisition of quinoa expression sequence data and unigene assembly. A total of 11.9G Illumina HiSeq 2000 RNA-Seq data (accession numbers: SRX257003 and SRX256971) and 2.1G Roche 454 EST data were obtained from NCBI's SRA database (http: / / www.ncbi.nlm.nih.gov / sra) (Accession number: SRX084791). After converting, cleaning, and filtering the original data, 19,571 unigenes were spliced using Trinity software, with a total base number of 80,448,006 bp, and the average length of each unigene was about 4kb. Among them, 16,854 sequences contained SSR loci.
[0018] 2) Identification of EST-SSR loci ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com