Method for identifying thelenota ananas by means of special peptide fragment group
A kind of plum ginseng, specific technology, applied in the biological field, can solve the problems of lack of technical level, deceiving consumers and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Sequence source and alignment information of specific polypeptides
[0037] 1. The source of the unique peptide ADIAEDSLK of plum ginseng
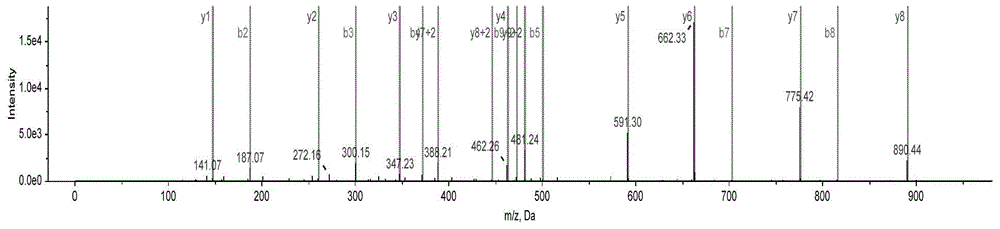
[0038] The unique polypeptide ADIAEDSLK of plum ginseng is derived from the protein tropomyosin [Apostichopusjaponicus]. Its Accesionnumber on NCBI is gi|302340969. The sequence is shown in SEQIDNO.5 below. Analysis shows that Glu at position 6 of ADIAEESLK has been mutated into Asp, and the resulting polypeptide ADIAEDSLK was compared by NCBIblast, and it was not found to be consistent with any other species:
[0039] SEQIDNO.5:
[0040] MDTIKKKLSQLKADKEKALDEKDVAEASMKEAMERVEQVNDENKELQTRIKQLETELDDTSEKLNTTVIKCEAAEKAQQTAEEEMANLQRKLQLTEEELSRSEERVADLQSKYTDIEQSSEENERQRKVLESRSAADDERMSELETQVMSSKTSLEDSDRKYDEASRKLTVTEEELARSEERSAAFESSLSQMKEELHQLHNNVKSLEAQEEKFTENEEMYEKKVRDLEDKLKVAEDRADIAEESLKSLKTSLDQLEDELMIEKEKVREMTEEMERTIQELNFEV
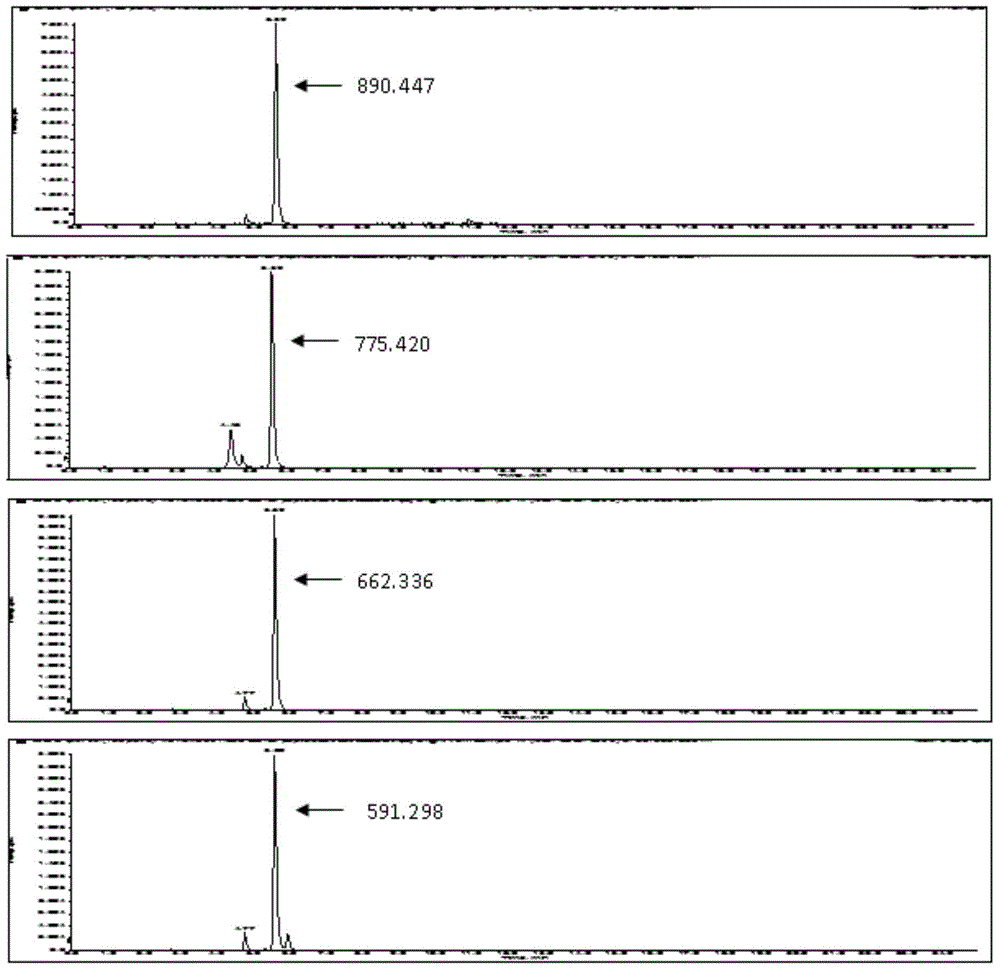
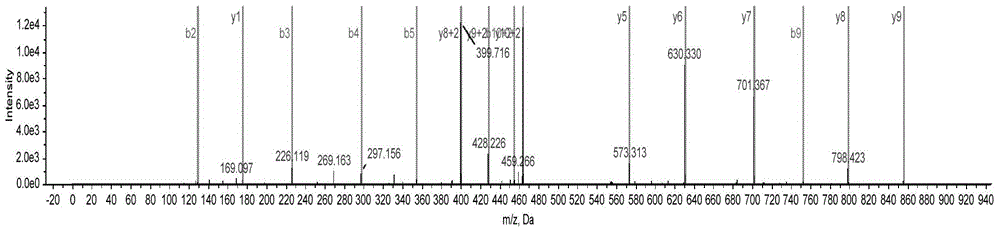
[0041] Adopt ABCIEX 5600 mass spectrometer (AB5600) detects the fragmentation and mass spectra of the target p...
Embodiment 2
[0066] Example 2, Sea cucumber sample processing and detection steps
[0067] The steps to analyze the sea cucumber sample to be tested include
[0068] (1) Sample pre-processing steps:
[0069] (1) Weigh 1g of sea cucumber sample and homogenize it into powder state, add 10mL protein extract (8M urea, 50mMNH 4 HCO 3 ) Extract protein by shaking, centrifuge at high speed and low temperature (20000r / min), take 200ul supernatant and transfer to 1ml EP tube;
[0070] (2) Take 2μL of 1mol / LDTT (0.154g dithiothreitol DTT dissolved in 1mL of deionized water) and add it to 100μL of the above protein solution to react at 37°C for 1 hour;
[0071] (3) Take 10 μL of freshly prepared IAA (accurately weigh 0.185 g of iodoacetamide IAA and dissolve in 1 mL of deionized water, and configure it into 1 mol / L IAA mother liquor for use. The configuration process needs to be protected from light. In the above reaction solution that has been cooled to room temperature, react at room temperature and avoid l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com