Liver cancer diagnosing reagent and kit using saliva as detection samples
A detection reagent and saliva detection technology, applied in the field of biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Saliva collection
[0038] Before saliva collection, the research individual should fast, drink, smoke, and clean the mouth for more than 2 hours.
[0039] In order to increase the amount of saliva collected, a sterile cotton swab was moistened with a 2% citric acid solution, and then the cotton swab was placed on the posterior side wall of one side of the individual’s tongue for about 5 seconds. After saliva was spit out, the cotton swab was placed on the other side. The posterior wall of the tongue. Collect repeatedly like this. The saliva collection volume needs to be more than 5ml, and the collection tube uses a 50ml sterile enzyme-free centrifuge tube. Centrifuge at 4°C at 3000g for 15min to take the supernatant from the upper layer to a 1.5ml Eppendoff tube, then centrifuge at 4°C at 12000g for 10min, then discard the precipitate and take the supernatant. All specimens were stored at -80℃ after processing.
Embodiment 2
[0040] Example 2 Extraction of total RNA from saliva
[0041] 1. Divide 1ml of saliva into two 1.5ml non-enzyme EP tubes equally, that is, each EP tube contains 500μl of saliva, then add 15μl of proteinase K (20mg / ml) to each EP tube and mix well , Incubate overnight in a 65 ℃ water bath. Because saliva contains more proteins such as digestive enzymes, which affect the subsequent extraction of salivary RNA, this method can remove the influence of salivary proteins on the experiment.
[0042] 2. Add 0.5ml of acid phenol and chloroform mixture (the main components are hydrochloric acid, phenol and chloroform) to the above saliva mixture in each of the two EP tubes.
[0043] Configuration of acid phenol chloroform
[0044] 2.1 The ratio of phenol to chloroform is 5:1
[0045] 2.2 Adjust the pH value with concentrated hydrochloric acid, the pH value is set at about 4.5, this pH value is most conducive to the extraction of RNA. Mix well.
[0046] 3. Shake well for 30 to 60 seconds after ad...
Embodiment 3
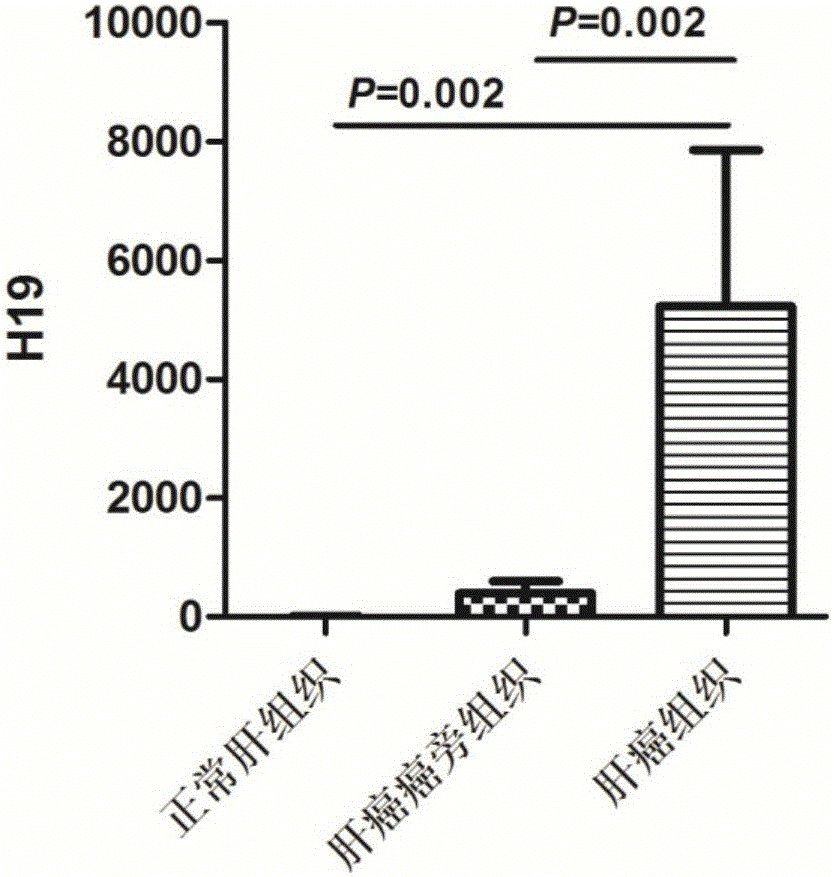
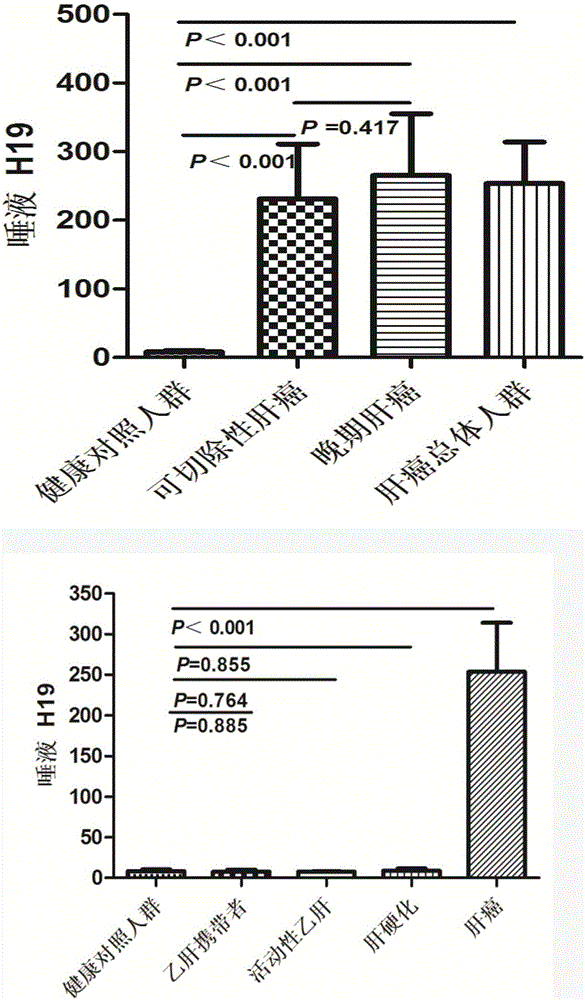
[0058] Example 3 Saliva lncRNAH19 reverse transcription and expression detection
[0059] 1. Reverse transcription reaction of saliva H19
[0060] Take 2μL RNA for reverse transcription. After reacting at 65°C for 5 min, immediately ice bath for 2 min.
[0061] Add 2μl 5×RTBuffer, 0.5μl EnzymeMix, 0.5μl PrimerMix (all Toyobo products, catalog number FSK-100) and 5μl DEPC-treated nuclease-free water to the above 2μL RNA, and then continue reverse transcription at 37℃ for 15min. ,98℃5min,4℃∞.
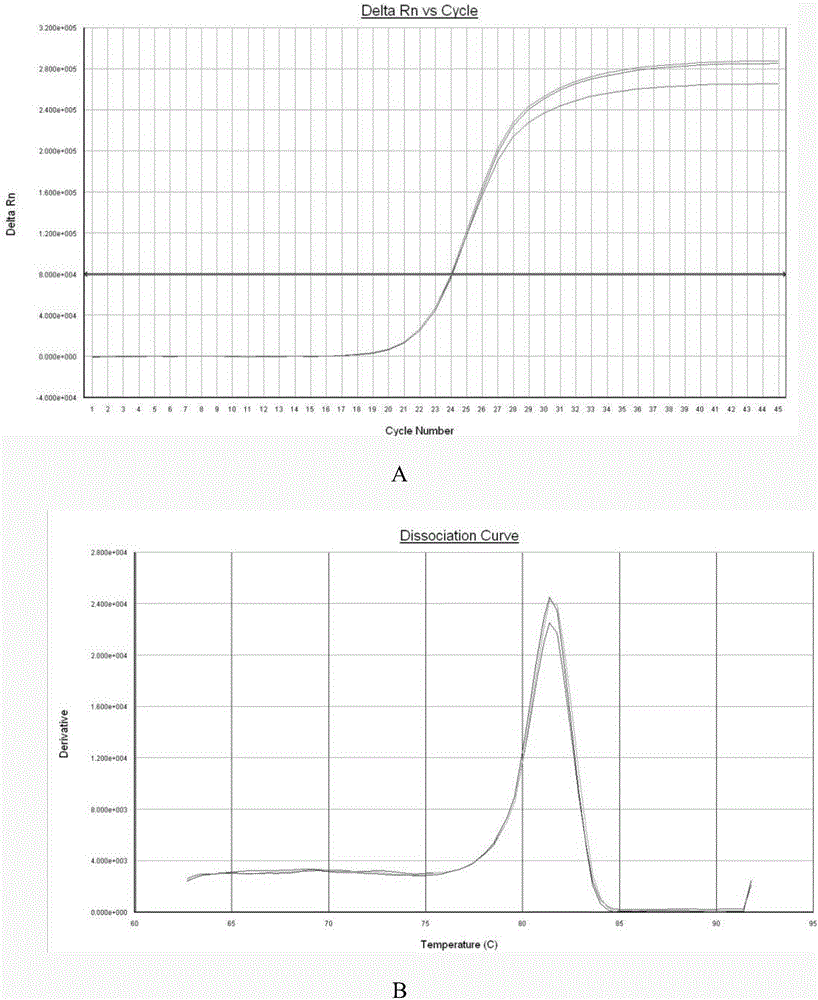
[0062] 2. Saliva RNAH19 expression detection (qPCR reaction)
[0063] Take 3μL of the reverse transcription product as a template, and use SYBRPremixExTaqⅡ Kit (TaKaRa Company, Item No. RB820A) for RT-qPCR detection. Add 9μL of 5×SYBRPremixBuffer, 0.8μL of upstream primer, 0.8μL of downstream primer, and 6.4 to 20μL reaction system. μL Enzyme-free water treated with DEPC.
[0064] Upstream primer sequence SEQIDNO.1: TGCTGCACTTTACAACCACTG
[0065] Downstream primer sequence SEQIDNO.2: ATGGTGTCTTTGA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com