CRISPR/Cas9 system for efficiently editing plant gene groups in fixed-point mode
A technology for encoding genes and plants, applied in angiosperms/flowering plants, plant products, genetic engineering, etc., can solve the problem of low editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1, construction of recombinant plasmid
[0061] 1. Construction of recombinant plasmid pYAO:Cas9
[0062] 1) Using the genomic DNA of wild-type Arabidopsis as a template, artificially synthesized pYAO-F: 5'-AA GTC GAC GATGGGAAATTCATTGAAAACCCT-3' (SalI restriction site is underlined) and pYAO-R: 5'-AA GTC GAC TCCTTTTCTTCTTCTCGTTGTTGT-3' (the underlined SalI restriction site) was used as a primer, and KOD-Plus-Neo was used for PCR amplification to obtain a double-stranded DNA molecule containing restriction endonuclease SalI at both the N-terminus and the C-terminus.
[0063] 2) After completing step 1), single-digest the double-stranded DNA molecule obtained in step 1) with restriction endonuclease SalI, and recover Fragment 1 of about 1024bp.
[0064] 3) The vector 35S-Cas9-SK was single-digested with the restriction endonuclease XhoI, and the vector backbone 1 of about 7493 bp was recovered.
[0065] 4) Ligate the fragment 1 with the vector backbone 1 t...
Embodiment 2
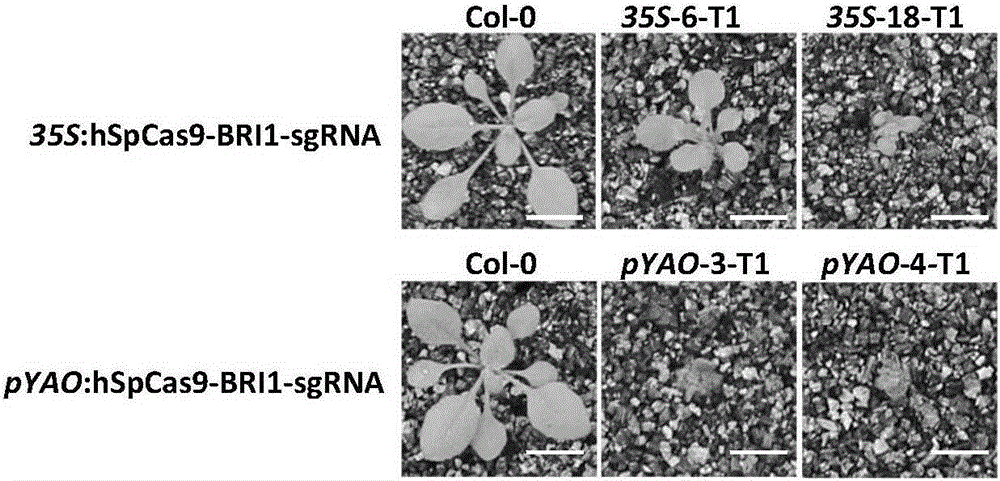
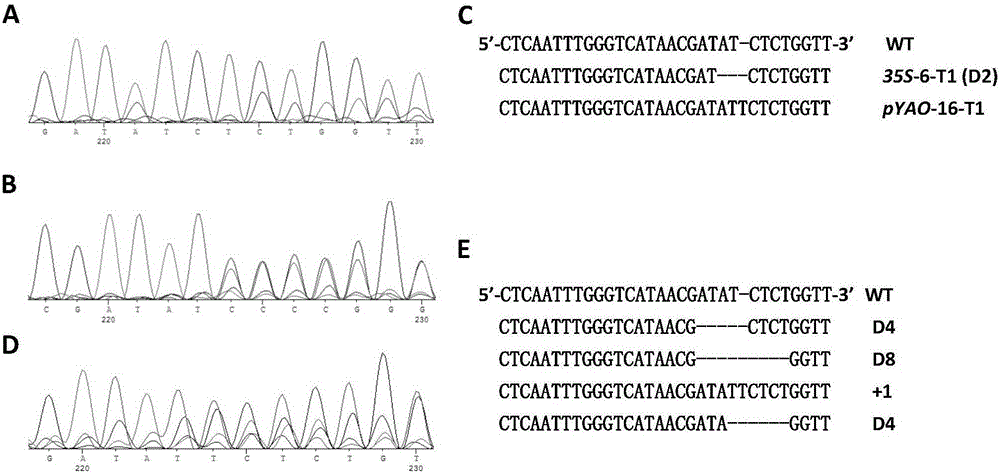
[0096] Example 2. Site-directed editing of Arabidopsis endogenous gene BRI1 by pYAO:Cas9 / AtU6-26-sgRNA system
[0097] 1. Design of target fragment BRI1-T1
[0098] Design the target fragment BRI1-T1, the target fragment BRI1-T1 is located on the target gene, and one strand in the double-stranded target fragment has the following structure: 5'-N X -NGG-3', N represents any one of A, G, C and T, X=20.
[0099] The nucleotide sequence of the target fragment BRI1-T1 is: 5'-TTGGGTCATAAC GATATC TC-3' (underlined is EcoRV enzyme recognition site).
[0100] 2. Construction of recombinant plasmid pYAO:hspCas9-BRI1-sgRNA
[0101] (1) Synthetic BRI1-T1F: 5'- ATTG TTGGGTCATAACGATATCTC-3' (the underlined part is sticky end) and BRI1-T1R: 5'- AAAC GAGATATCGTTATGACCCAA-3' (the underlined part is the cohesive end), BRI1-T1F and BRI1-T1R are all single-stranded DNA molecules.
[0102] (2) Mix BRI1-T1F and BRI1-T1R at a molar ratio of 1:1 and anneal (annealing procedure: 95°C, 5min, n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com