A Fluorescence Quantitative PCR Detection Method of Xijiang Virus Sybr Green I
A fluorescence quantitative and Xijiang virus technology, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of no patent publication of Xijiang virus detection technology, and achieve improved sensitivity, high sensitivity, and simple operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0028] Specific embodiment one: a kind of Xijiang virus SYBR Green I fluorescent quantitative PCR detection method of the present embodiment, it is carried out according to the following steps:
[0029] 1. Design of XRV SYBR Green Ⅰ fluorescent quantitative PCR primers
[0030] According to the sequence of the XRV S1 gene, sequence comparison was performed by the software clustalx1.81 to determine the specific sequence region of the XRV gene, and fluorescent quantitative primers specific for XRV were designed in this region; the primer sequences are as follows:
[0031] XRV-F: 5'CTAACTCCGCTGACACGA 3';
[0032] XRV-R: 5'CAAACACTATTGACGCACAT 3';
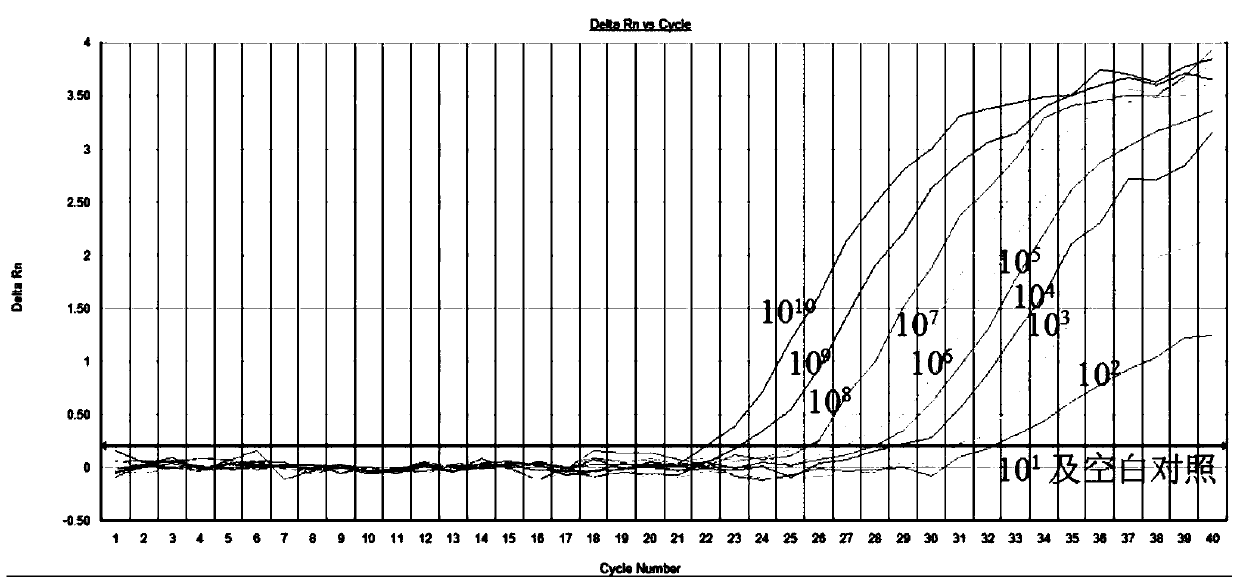
[0033] 2. Construction of standard curve
[0034] According to the primers designed in step 1, the full-length XRV S1 gene cDNA was used as a template to carry out fluorescent quantitative amplification, and draw a fluorescent quantitative standard curve;
[0035] 3. Experimental establishment standards
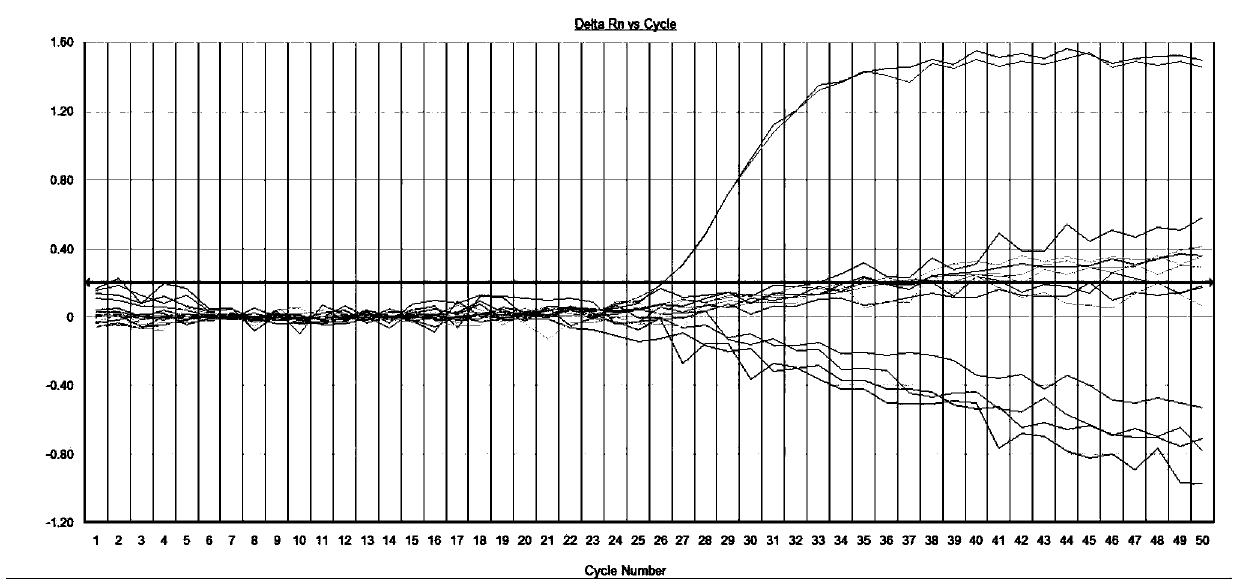
[0036] If the blank contro...
specific Embodiment approach 2
[0039] Embodiment 2: The difference between this embodiment and Embodiment 1 is that the reaction system of fluorescent quantitative PCR is 25 μl, and the specific components are:
[0040]
[0041] The fluorescent quantitative PCR reaction program was as follows: pre-denaturation at 95°C for 2 min, denaturation at 95°C for 10 sec, annealing and extension at 60°C for 20 sec, and fluorescence signal collection, a total of 40 cycles.
[0042] Others are the same as in the first embodiment.
specific Embodiment approach 3
[0043] Embodiment 3: This embodiment is different from Embodiment 1 in that: the full-length S1 gene cDNA of XRV is obtained by reverse transcription of the full-length S1 gene DNA of XRV. Others are the same as in the first embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com