Selenium-rich candida glabrata strain FXY-4 as well as cultivation method and application of selenium-rich candida glabrata strain FXY-4 serving as additive for fish feed
A technology of Candida glabrata and FXY-4, which is applied in the field of microorganisms, achieves good application prospects, saves breeding costs and improves economic benefits.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Candida glabrata FXY-4, which can be obtained as follows:
[0021] Isolation of the original strain:
[0022] Dilute the pickle juice sample and spread it on the plate of YEPD medium, cultivate it at 28°C, observe the growth state of the colony at regular intervals, and pick a single colony and transfer it to the YEPD slant for storage. After preliminary screening, the following yeasts are obtained: Strains: FXY-1, FXY-3, FXY-4, FXY-5, FXY-7, FXY-10, FXY-11, FXY-19, FXY-21, FXY-23, FXY-24, FXY-25, FXY-38, FXY-42, FXY-43, FXY-44, FXY-46, FXY-47, FXY-49, FXY-50, FXY-53, FXY-55, FXY-56.
[0023] containing Na 2 SeO 3 (final concentration is 0.5g / L) in the YEPD substratum that carries out the screening of selenium-enriched yeast, then the yeast that preliminary screening obtains is cultivated with YEPD substratum, and every strain bacterium is divided into blank group and experimental group and cultivates, Add Na to the experimental group 2 SeO 3 (final concentration ...
Embodiment 2
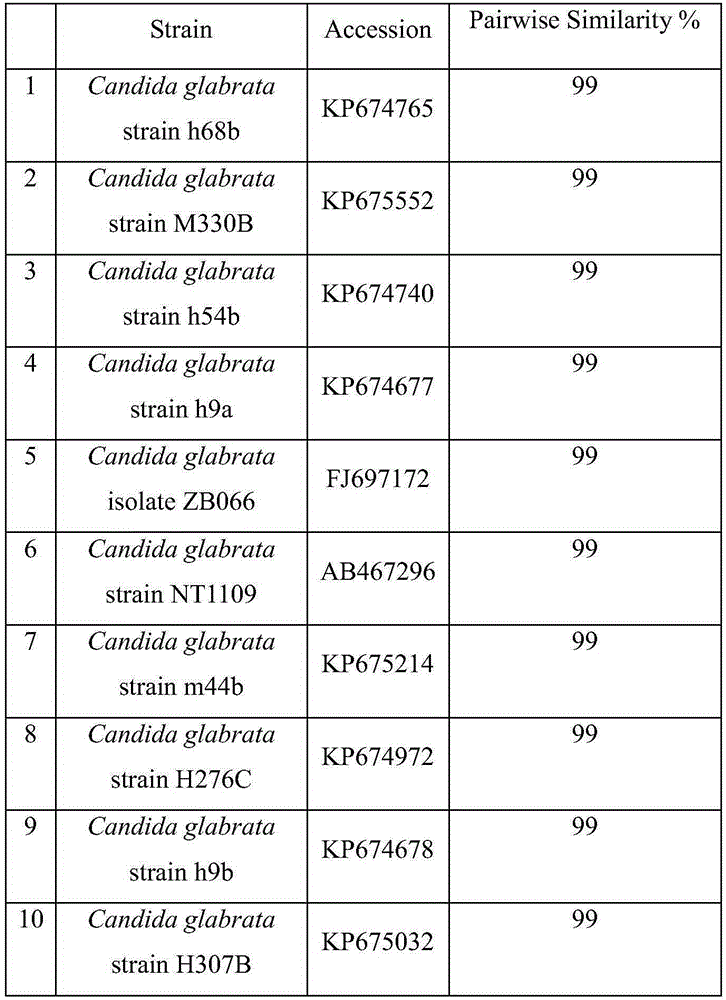
[0025] Yeast strain FXY-4 (deposit number: CCTCCNO: M2015664) 18SITSrRNA gene sequence analysis:
[0026] (1) Extraction of chromosomal DNA (small amount):
[0027] 1. Take 1.5mL bacterial liquid in a 1.5mL Eppendorf tube and centrifuge at 12000r / min for 5min;
[0028] 2. Discard the supernatant, resuspend the pellet in 900 μL phosphate buffered solution (PBS), and centrifuge at 12000 r / min at 4 °C for 5 min;
[0029] 3. Discard the supernatant, add 300TE and 200μL 10mg / mL lysozyme, blow and aspirate to mix, incubate at 37°C for 1 hour, and mix by inverting every 15 minutes;
[0030] 4. Add 600TENS lysate (200mmol / LNaCl, 100mmol / LTril-HClpH8.0, 2.0%SDS, 50mmol / LEDTA, 0.5%TritonX-100) and 20mg / mL proteinase K10μL to the pellet, mix upside down, and incubate at 55°C 1h, invert and mix once every 15min;
[0031] 5. Centrifuge at 12000r / min at 5.4°C for 5min, and take the supernatant;
[0032]6. Add an equal volume of P:C:I (25:24:1) to the supernatant, mix thoroughly, and cen...
Embodiment 3
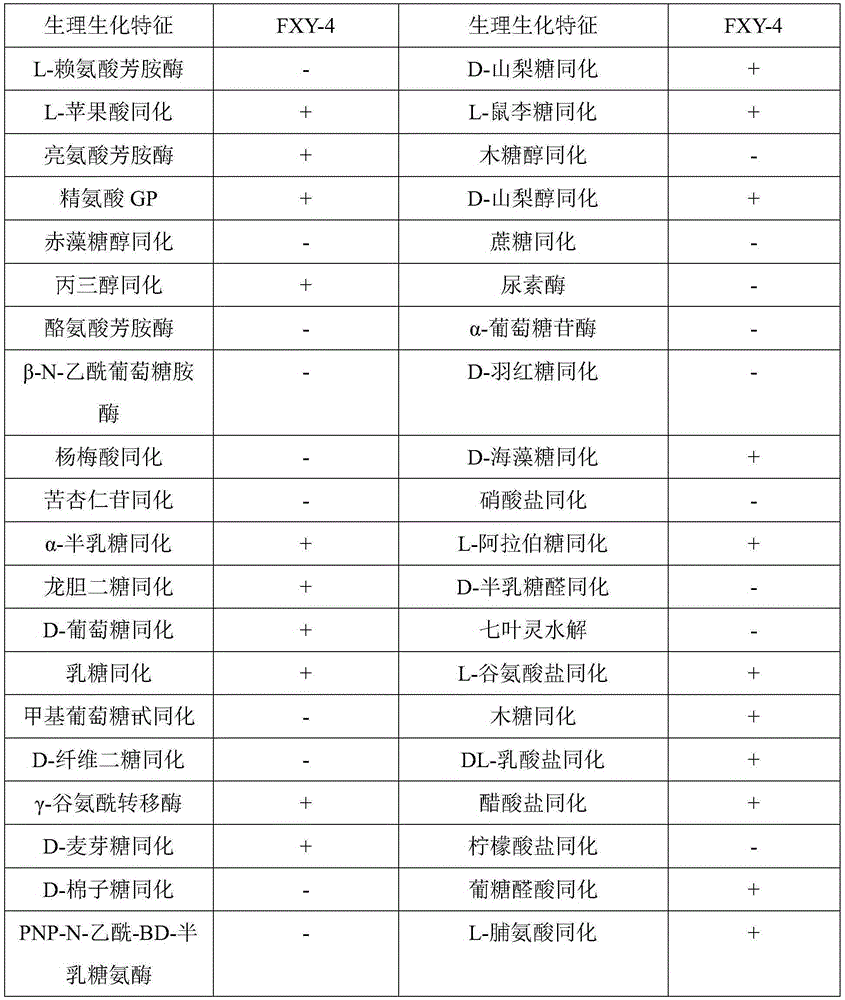
[0060] Yeast FXY4 (Deposit number: CCTCCNO: M2015664), BioMérieux VITEK2 automatic identification analysis
[0061] Pick the bacterial lawn of the strain to be tested and inoculate it on the YEPD plate, cultivate it to the logarithmic growth phase at 30°C, dip the Vitek liquid in the kit with a sterilized cotton swab, scrape off the bacterial lawn on the plate, and then dissolve it in 1.8ml of Vitek liquid. Shake and mix well; use a turbidimeter to adjust the turbidity to the specified range (2MacF); then use a pipette gun to add the bacterial suspension to the Vitek identification plate, and the bacterial solution should fill the identification well; then incubate at 30°C for 24 to 48 hours Take out the identification plate, and use the BioMérieux VITEK2 automatic identification analyzer to read and analyze the data of the identification plate. The results are shown in Table 3. The physiological and biochemical results are the same as the typical physiological and biochemical ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com