Application of novel fructosidase coding gene
A technology of fructosidase and fructanase, applied in the direction of glycosylase, enzyme, hydrolase, etc., can solve the problem of insufficient types of fructosidase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Determination of the position of β-fructofuranosidase in Lactobacillus plantarum and extraction of crude enzyme
[0044] After culturing Lactobacillus plantarum (Lactobacillus plantarum) ST-III at 37° C. for 24 h, the supernatant and cells were collected by centrifugation (4° C., 12,000×g, 10 min). After the supernatant was sterilized by 0.45 μm membrane filtration, it was concentrated to 1 / 20 of the original volume by ultrafiltration through a 25 kDa ultrafiltration membrane, which was the supernatant of the fermentation broth. After the cells were washed twice by centrifugation with 50 mMPBS (pH=6.0), lysozyme was added to 2 mg / mL, and then acted at 37° C. for 1 h. Sonicate intermittently in an ice bath for 8 minutes (400W, ultrasonic 5s, intermittent 5s), and then centrifuge (4°C, 12,000×g, 10min) to collect the supernatant and precipitate respectively. The pellet was resuspended in an equal volume of the above-mentioned PBS, which was the broken cell pell...
Embodiment 2
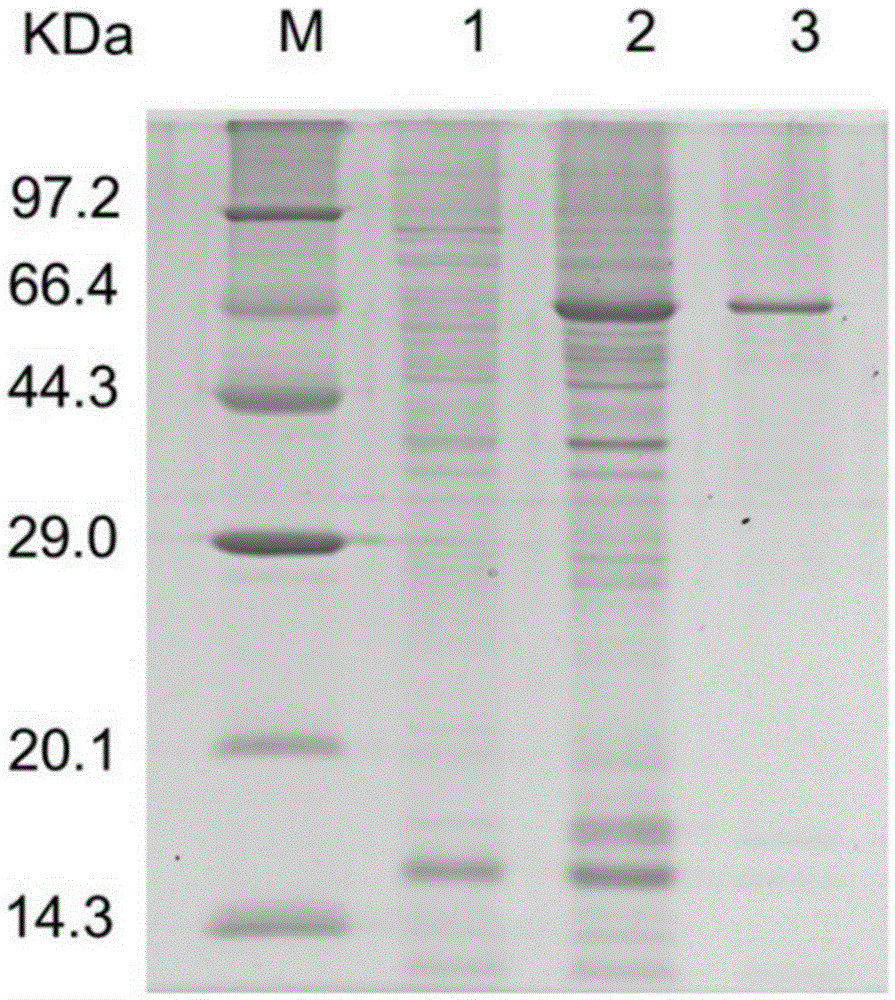
[0049] Cloning expression and property determination of embodiment 2 fructosidase
[0050] 1. Construction of recombinant expression plasmids
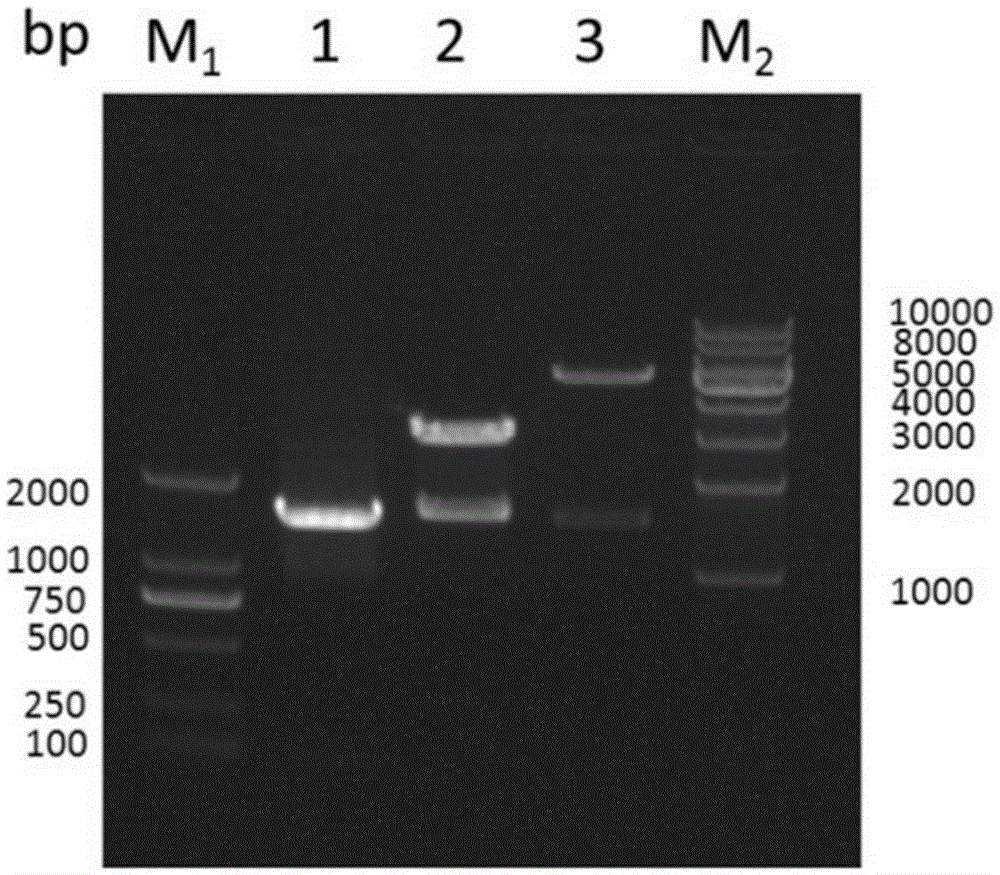
[0051] Using SEQ ID No. 3: 5'-GGAATTCCATATGGAGCTCATCGTCATGATATGGAAT-3' and SEQ ID No. 4: 5'-CCGCTCGAGCCGCTAGCTTCGCAACATC-3' as primers, and Lactobacillus plantarum ST-III genome as template, PCR reaction was carried out. The PCR reaction system is 20.0μL, and the final concentration of each reaction is: 1×PCRbuffer, 1mmol / L Mg 2+ , the dNTP of 0.2mmol / L, the upstream and downstream primers (SEQIDNo.3, SEQIDNo.4) of 0.4μmol / L, the TaqDNA polymerase of 0.1U / μL and the DNA template of 2.0ng / μL, use ddH 2 O to make up to 20.0 μL. The PCR reaction parameters were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 90 s, 35 cycles; extension at 72°C for 7 min. Amplified products were detected by 1% agarose gel electrophoresis. Ligate the purified target gene fragments to ...
Embodiment 3
[0064] The determination of embodiment 3β-fructofuranosidase properties
[0065] 1. Determination of β-fructofuranosidase enzyme activity
[0066] The β-fructofuranosidase assay uses kestose as the substrate, and the system includes: 50 μL 20 mM kestose, 50 μL 50 mMPBS (pH 6.0), 15 μL enzyme solution (11 μg / mL), and the volume is made up to 200 μL with water. Mix well at 37°C for about 10 minutes, and then in a water bath at 100°C for 5 minutes. The fructose produced was detected by a fructose assay kit (BioVision, USA). Definition of β-fructofuranosidase enzyme activity: Under the conditions of 37°C and pH 6.0, 1 μmol of fructose produced by hydrolyzing kestose per minute is defined as a unit. And the specific enzyme activity of the enzyme was calculated by measuring the protein content.
[0067] 2. Determination of optimum pH of β-fructofuranosidase
[0068] Prepare different pH buffers (pH3.0–8.0, 0.2MNa 2 HPO 4 / 0.1M citrate buffer; pH8.5–9.0, 50mM Tris–HCl buffer) t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com