Recombinant expression vector of FGFR3 mutant as well as construction method and application of recombinant expression vector
A technology for FGFR3 and expression vectors, which is applied in the field of recombinant expression vectors of FGFR3 mutants and its construction, can solve the problems of the construction method and application of FGFR3 mutant recombinant expression vectors, lack of effective molecular markers, hysteresis, etc., and achieve good results. Application value, high transfection efficiency, and stable expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Construction of recombinant expression vector
[0051] 1. Construction of pWPI.1-FGFR3IIIc-IRES2-EGFP, pWPI.1-FGFR3AT-I-IRES2-EGFP and pWPI.1-FGFR3Δ7-9-IRES2-EGFP vector construction and pcDNA3.0-FLAG-FGFR3, pcDNA3.0- HA-FGFR3, pcDNA3.0-FLAG-FGF1, pcDNA3.0-HA-FGF1, pcDNA3.0-FLAG-FGF2, pcDNA3.0-HA-FGF2 vector construction (the above vectors are all FGFR3 wild type IIIc, FGFR3 mutant ATI and mutant Δ7-9-related vector construction, as well as the vector construction of the tag protein in some experiments)
[0052] 1. PCR primer design and target gene amplification
[0053] The primers were designed by Primer5.0 software, the sense strand: FGFR3-F5ˊ-gacGGATCCatgggcgcccctgcctgcgccctc-3ˊ, as shown in SEQ ID NO.2; the antisense strand: FGFR3-R5ˊ-gacACTAGTtcacgtccgcgagcccccactgct-3ˊ, as shown in SEQIDNO. Synthesized by bioengineering company. The 5'end restriction site of the sense strand is BamHI, and the 5'end restriction site of the antisense strand is SpeI. KODplus ...
Embodiment 2
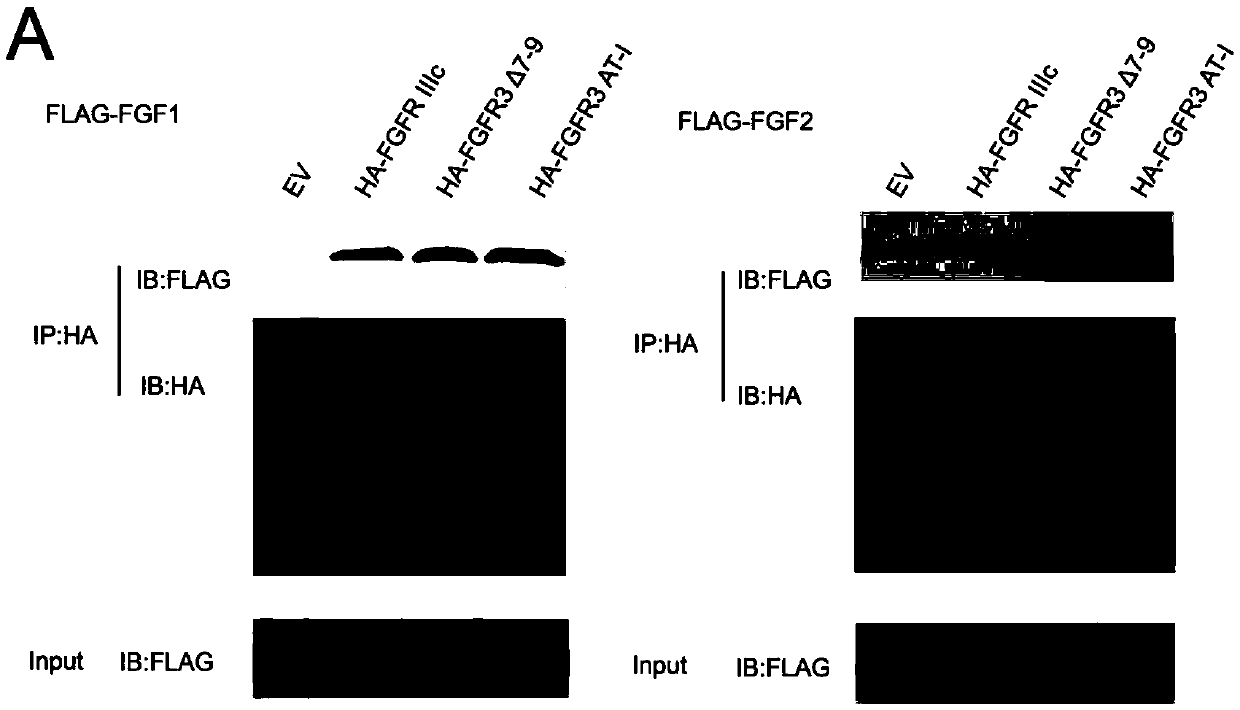
[0183] Example 2. Identification of the affinity of FGF1 / FGF2 and FGFR3 and their mutants
[0184] 1. Co-immunoprecipitation to identify the binding ability of FLAG-FGF1, FLAG-FGF2, HA-FGFR3 and its mutants
[0185] 1. Routinely cultivate 293T cells co-transfected with HA-FGFR3 and FLAG-FGF1 for two days, harvest the cells, add an appropriate amount of RIPA lysis buffer (millipore) containing protease inhibitors (roche, protease inhibitor tablets), and lyse on ice or at 4°C for 30 minutes , Then centrifuge at 12,000g for 30min and take the supernatant;
[0186] 2. Take a small amount of lysate (usually 5% of the total amount) as input for Westernblot analysis, add 1-2μg HAprobe (santacruz) to the remaining lysate, incubate at 4°C for 2h and then add 20-40μl proteinA / G- Beads (santacruz), incubate overnight with slow shaking at 4°C. This step is mainly to immunoprecipitate HA-FGFR3;
[0187] 3. After the immunoprecipitation reaction, centrifuge at 3,000g at 4°C for 5 minutes, and cent...
Embodiment 3
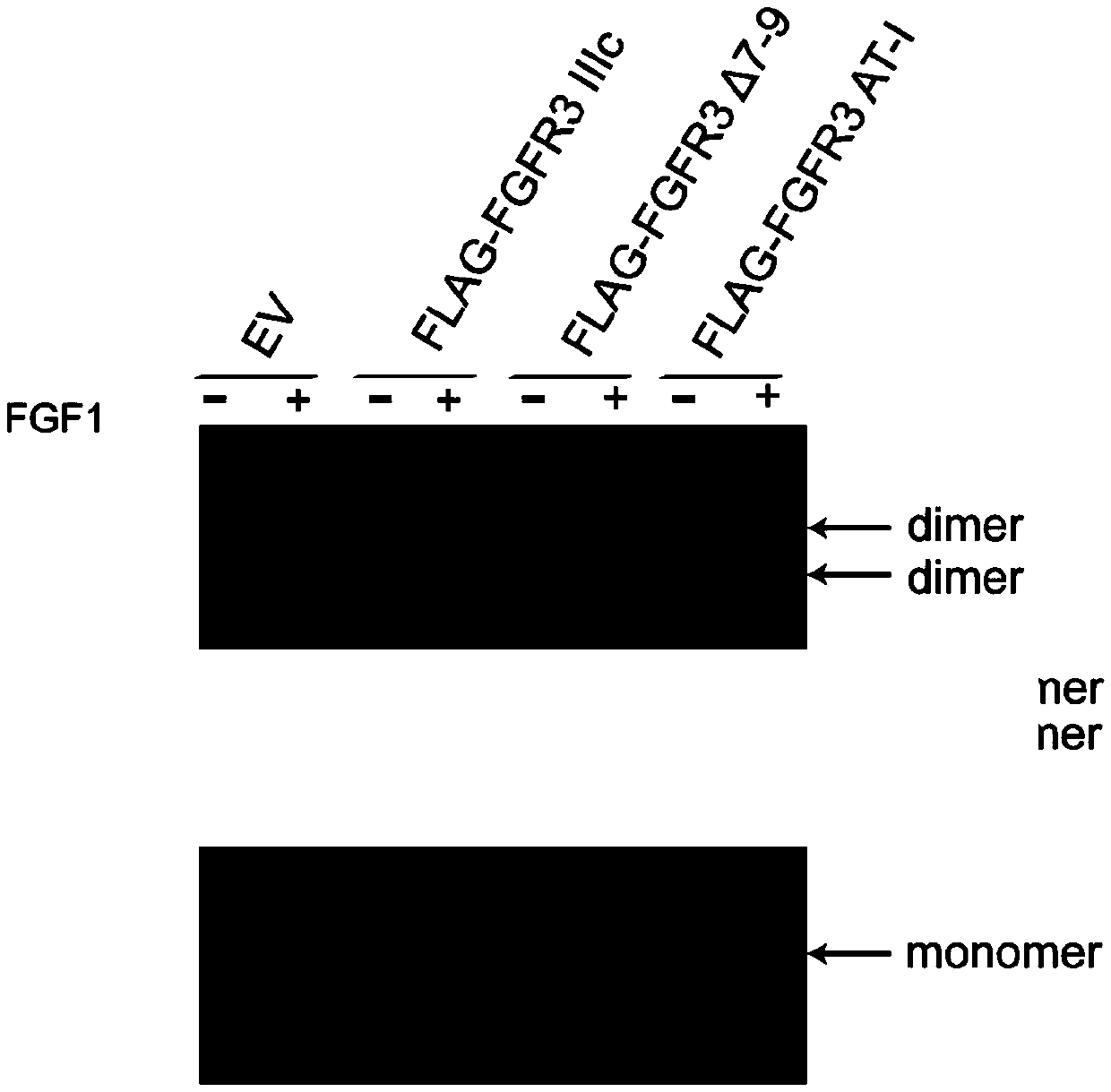
[0212] Example 3 Identification of the ability of FGFR3 and its mutants to form dimers
[0213] 1. 293T cells were transiently transferred with pcDNA3.0 empty, pcDNA3.0-FLAG-FGFR3IIIc, pcDNA3.0-FLAG-FGFR3Δ7-9 and pcDNA3.0-FLAG-FGFR3AT-I, divided into two groups, one group did not add FGF1, The other group was routinely cultured with medium containing 10ng / ml FGF1 for 2 days. Add an appropriate amount of RIPA lysis buffer (millipore) containing protease inhibitors (roche, protease inhibitor tablets), lyse on ice or 4°C for 30 min, then centrifuge at 12,000 g for 30 min and take the supernatant.
[0214] 2. Load the sample to non-denaturing polyacrylamide gel electrophoresis (Native-PAGE) without SDS, and perform WesternBlotting.
[0215] by Figure 7 It can be seen that FLAG-FGFR3IIIc hardly forms dimers without FGF1 stimulation, but can form dimers under FGF1 stimulation; and FLAG-FGFR3Δ7-9 can form dimers regardless of whether FGF1 is stimulated or not; Regardless of FGF1 stimulat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com