Gene chip for detecting gamma-proteobacteria communities in marine environment
A technology of gene chip and marine environment, applied in the field of gene chip for detecting gamma-proteobacteria community in marine environment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
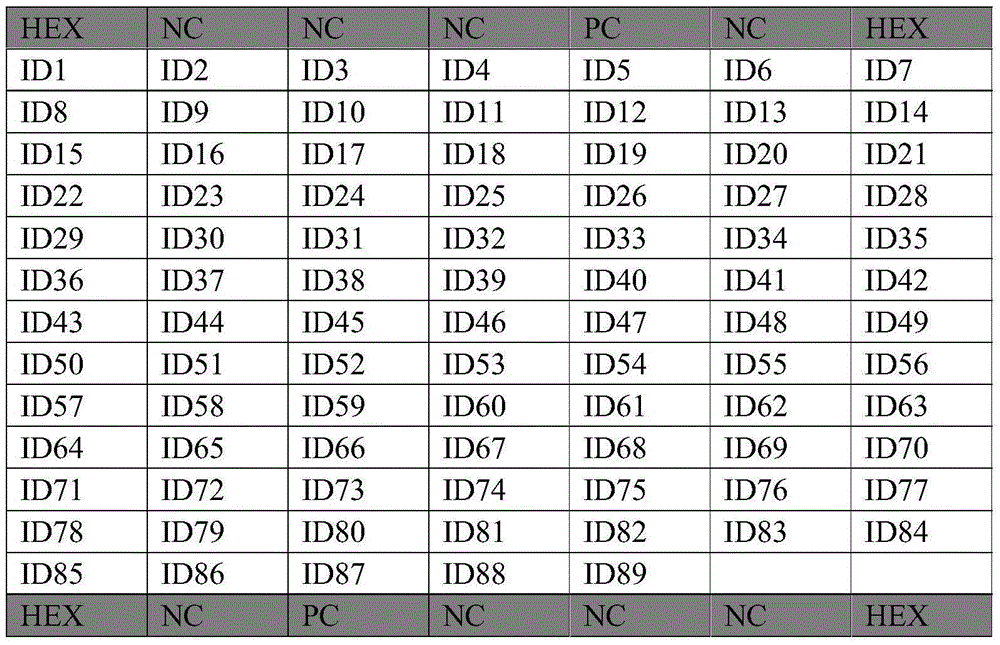
[0047] Example 1: Design of probes for gene chips for gamma-proteobacteria community detection
[0048] High-throughput sequencing technology and clone library technology were used to obtain the community information of γ-proteobacteria in seawater of East China Sea. According to the 16S rRNA gene sequence information of the γ-proteobacteria community, the 31 main γ-proteobacteriaceae (Aeromonadaceae, Alcanivoracaceae, Alteromonadaceae, Chromatiaceae, Colwelliaceae, Coxiellaceae, Ectothiorhodospiracea, Enterobacteriaceae, Thiotrichaceae, Xanthomonadaceae, Hahellaceae, Hahellaceae, Halomonadaceae、Idiomarinaceae、Legionellaceae、Litoricolaceae、Oceanospirillaceae、Zooshikella、Vibrionaceae、Moraxellaceae、Thiotrichales_incertae_sedis、Pasteurellaceae、Piscirickettsiaceae、Pseudoalteromonadaceae、Pseudomonadaceae、Psychromonadaceae、Salinisphaeraceae、Shewanellaceae、Sinobacteraceae、Ferrimonadaceae、Francisellaceae、Methylococcaceae)进行探针设计,并在BLAST中进行 To verify, obtain the probe sequence used to pr...
Embodiment 2
[0085] Embodiment 2: Specific detection of the gamma-proteobacteria colony detection gene chip of the present invention
[0086] The representative clone Francisellaphilomiragia1-5 of Francisellaceae, a seawater dominant family Francisellaceae in γ-proteobacteria, was selected to detect the specificity of the gene chip for γ-proteobacteria community detection.
[0087] ① DNA extraction: Plasmid DNA was extracted with a plasmid extraction kit.
[0088] ②Amplification of 16SrRNA gene: The 16SrRNA gene was linearly amplified using the cloning vector pEASY-T1 universal primers M13F (GTAAAACGACGGCCAGT) and M13R (GTCCTTTGTCGATACTG). The amplification program was 94°C for 5min, 25 cycles (94°C for 30s, 55°C for 30s, 72°C for 1min30s), and 72°C for 10min.
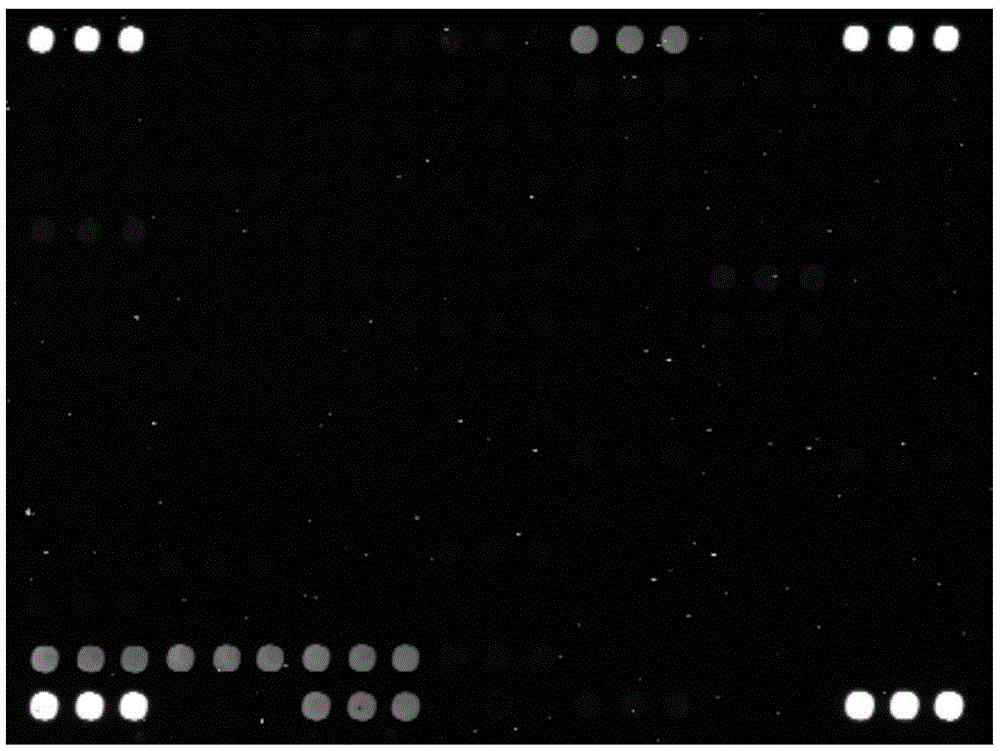
[0089] ③Fluorescent labeling: Use fluorescently labeled random primers (Cy3-NNNNNNNNNN) and Klenow enzyme to label the 16SrRNA gene amplification products. 1.5h at 37°C, 10min at 70°C.
[0090] ④ Hybridization: 15 μL of fluoresc...
Embodiment 3
[0092] Embodiment 3: Utilize the chip to detect the gamma-proteobacteria species in the seawater sample
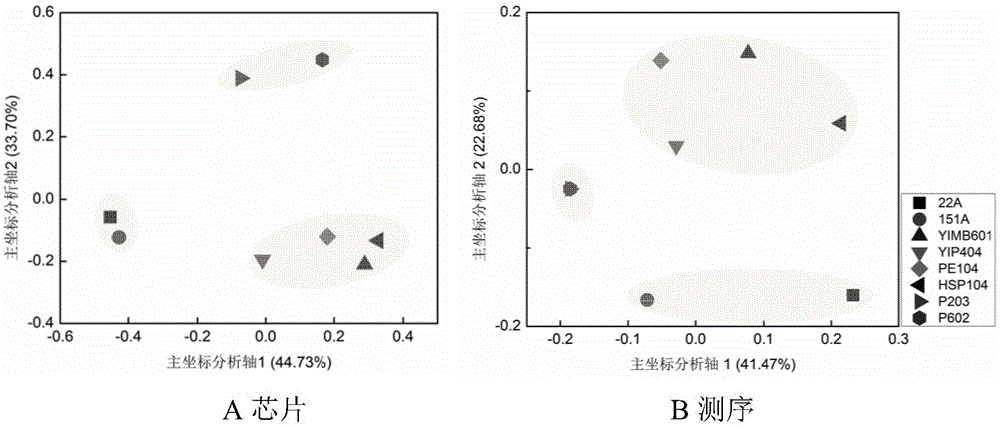
[0093] The seawater sample 22A was hybridized with the marine γ-proteobacteria community detection gene chip of the present invention, the specific method is as in Example 2. And compared with the results of high-throughput sequencing. The results are shown in Table 1.
[0094] Table 1 Detection results of γ-proteobacteria in seawater sample 22A
[0095]
[0096]
[0097] Note: + means detected, - means not detected.
[0098] 基因芯片检测结果表明海水样品22A中含有zooshikella、Xanthomonadaceae、Vibrionaceae、Thiotrichales_incertae_sedis、Thiotrichaceae、Sinobacteraceae、Shewanellaceae、Psychromonadaceae、Pseudomonadaceae、Pseudoalteromonadaceae、Piscirickettsiaceae、Pasteurellaceae、Oceanospirillaceae、Moraxellaceae、Methylococcaceae、Litoricolaceae、Legionellaceae、Idiomarinaceae、Halomonadaceae、Hahellaceae、 Francisellaceae, Enterobacteriaceae, Ectothiorhodospiraceae, Coxiellaceae, Colwelliaceae, Ch...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com