Diagnosis and treatment markers for hysteromyoma
A technology for uterine fibroids and products, which can be applied in gene therapy, biological testing, anti-tumor drugs, etc., and can solve the problems of early diagnosis of uterine fibroids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
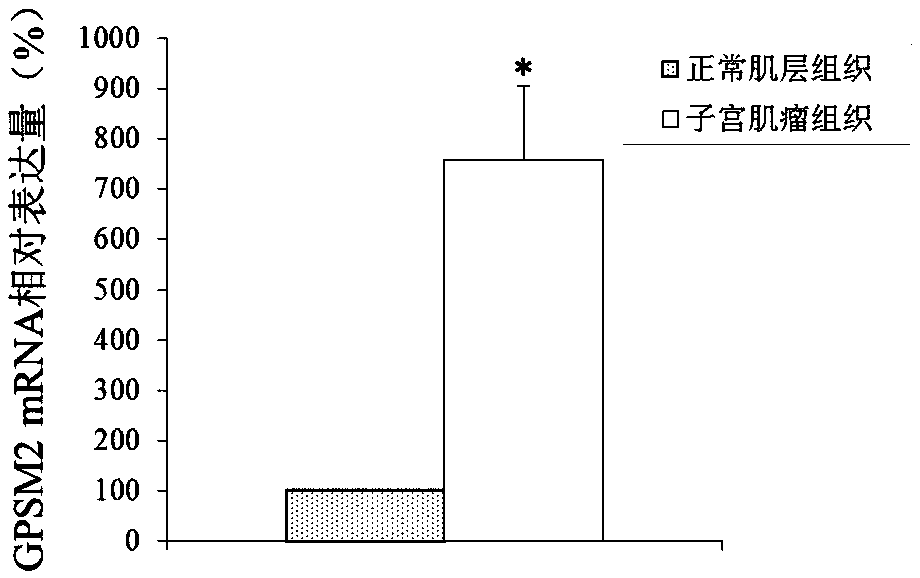
[0072] Differential expression of embodiment 1GPSM2 gene
[0073] 1. Experimental materials:
[0074] Uterine fibroid tissue and adjacent normal muscle layer tissue were aseptically collected from patients undergoing total hysterectomy for uterine fibroids. The patient had not received hormone therapy within 3 months before operation, and all patients were pathologically diagnosed as uterine fibroids after operation. , the experimental materials were taken from 40 patients with uterine fibroids, and the patients were between 25 and 45 years old.
[0076] Relying on the technical service of the gene chip analysis platform provided by Biobio, the AFFX human genome expression profile chip (AFFXHumanGenomeU133Plus2.0Array, Affimetrix Company) was selected to complete the gene chip screening analysis of tissue RNA samples, its working principle and the entire operation process Referring to the company's conventional gene chip analysis method, the m...
Embodiment 2
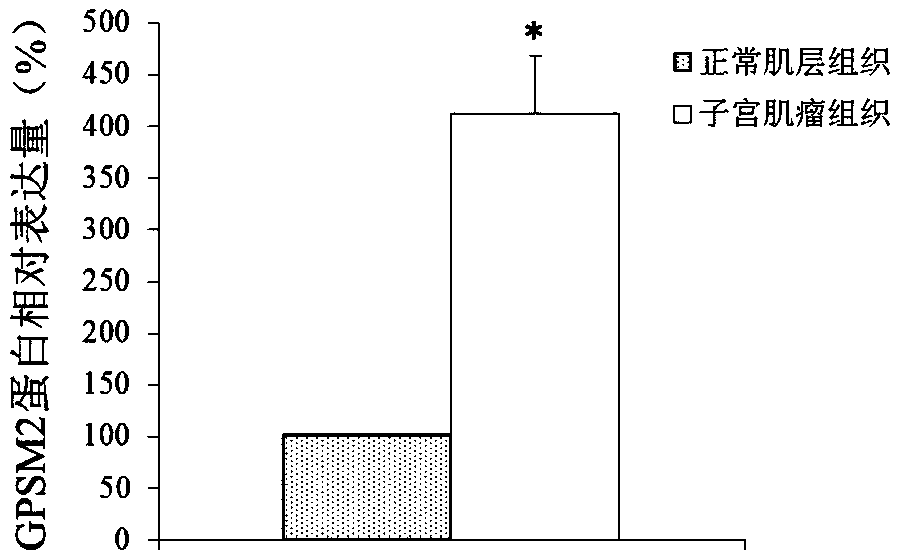
[0085] Differential expression of embodiment 2GPSM2 protein
[0086] 1. The research object is the same as in Example 1.
[0087] 2. Extraction of total tissue protein
[0088] Follow the instructions of the EpiQuik Tissue / Cell Total Protein Extraction Kit for protein extraction.
[0089] 3. Western blot detection
[0090] The total protein was quantified by the Brandford method, take an appropriate amount and mix it with the sample buffer, boil for 5 minutes, and cool for 5 minutes; take 30pg of protein and load it on a prepared 15% polyacrylamide gel for electrophoresis, start with a constant voltage of 80V, and see the Marker Increase to 120V; take out the gel after electrophoresis, use Bio.Rad semi-dry transfer system to transfer at 100V for 50min; after the transfer is completed, wash once with 1xPBS, immerse in blocking solution, 40C overnight; pour off blocking solution, add Western wash Wash with the membrane washing solution for 5-10 minutes, add the primary antibo...
Embodiment 3
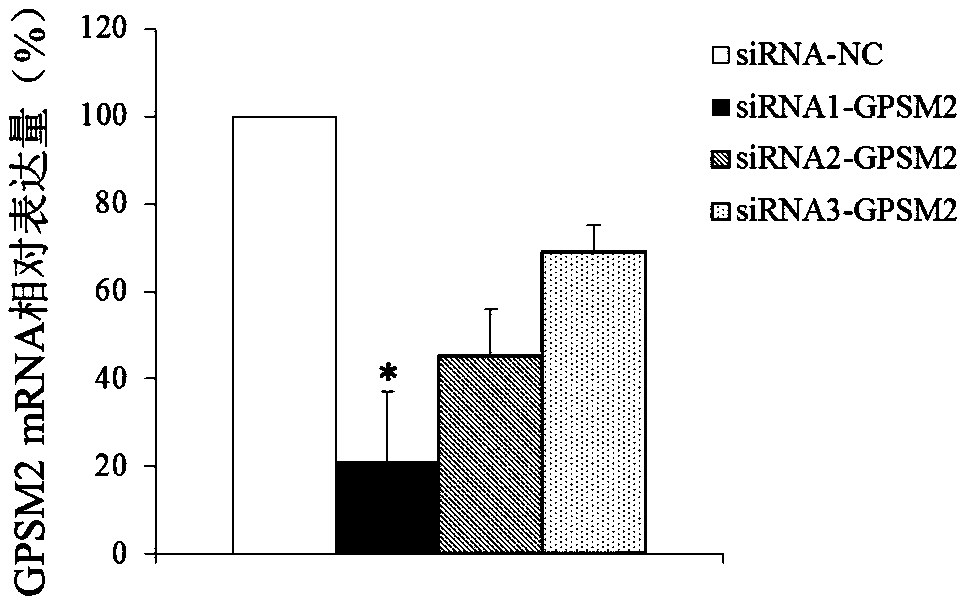
[0095] Embodiment 3 suppresses GPSM2 gene expression
[0096] 1. siRNA design and synthesis
[0097] siRNA sequence against GPSM2:
[0098] siRNA1-GPSM2:
[0099] The sense strand is 5'-UCACUAAUGAUAGGUUUUCCU-3' (SEQ ID NO.7);
[0100] The antisense strand is 5'-GAAAACCUAUCAUUAGUGACU-3' (SEQ ID NO.8),
[0101] siRNA2-GPSM2:
[0102] The sense strand is 5'-AGUAAAGUAUAUGUAUUUCCA-3' (SEQ ID NO.9);
[0103] The antisense strand is 5'-GAAAUACAUAUACUUUACUUC-3' (SEQ ID NO.10),
[0104] siRNA3-GPSM2:
[0105] The sense strand is 5'-AUUAUGGAGUUAUUUUGUGCUG-3' (SEQ ID NO.11);
[0106] The antisense strand is 5'-GCACAAAUAACUCCAUAAUGU-3' (SEQ ID NO.12)
[0107] Negative control siRNA sequence (siRNA-NC):
[0108] The sense strand is 5'-CGUACGCGGAAUACUUCGA-3' (SEQ ID NO.13);
[0109] The antisense strand is 5'-UCGAAGUAUUCCGCGUACG-3' (SEQ ID NO.14).
[0110] 2. Culture and transfection of uterine leiomyoma cells
[0111] After surgical resection of uterine fibroids, 1 cm of each u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com