Method for detecting variation of copy numbers of genomes
A technology of copy number variation and genome, applied in the field of detection of genome copy number variation, can solve the problems of unstable probe hybridization efficiency, PCR pollution, low resolution, etc. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
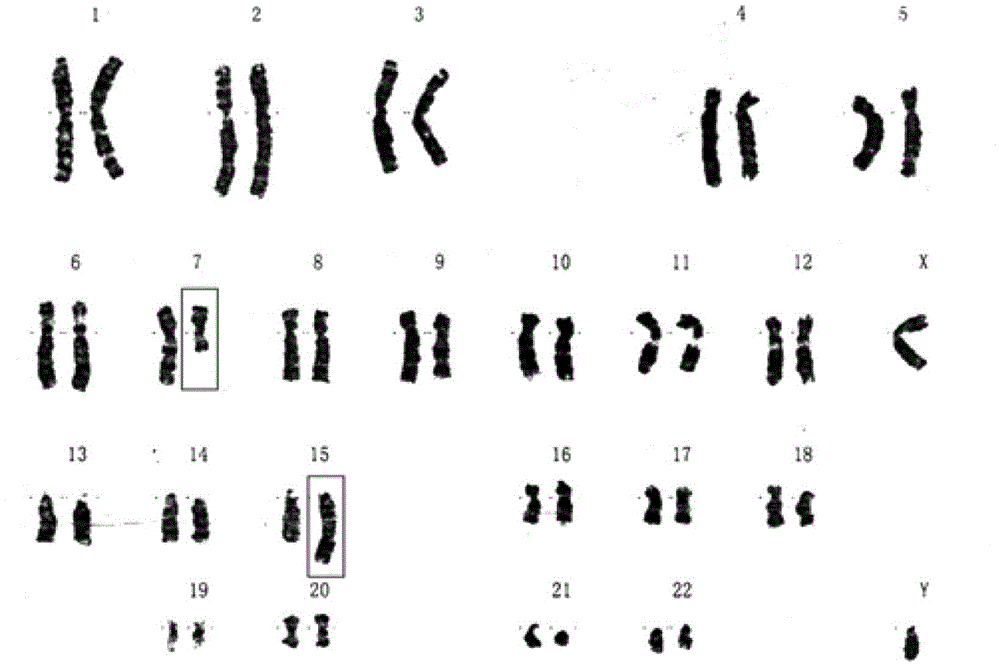
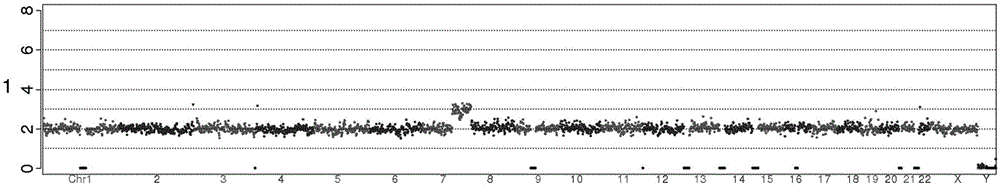
[0038] In this embodiment, the copy number variation detection is performed on the cell samples of two generations, and the detection results are compared with the paternal karyotype results.
[0039] 1. Sequencing
[0040] In this embodiment, for the detection of a sample containing a trace amount of nucleic acid, firstly, single-cell whole-genome amplification is performed. Single cell expansion using MALBAC of Yikang Gene Technology Co., Ltd. SingleCellWholeGenomeAmplificationKit, the single cell is a sample containing a small amount of nucleic acid, such as free single cells in blood, urine, and saliva.
[0041] The amplified samples were purified, the library was constructed, and sequenced on the machine. The on-machine sequencing used the HiSeq2500 high-throughput sequencing platform of Illumina, and operated according to the instructions provided by Illumina. The sequencing type is single-end (SingleEnd) sequencing, the sequencing length is 50bp, and the sequencing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com