Polypeptides capable of realizing targeted editing of GS gene, and proteins thereof
A targeting and gene technology, applied in the field of genetic engineering, can solve the problems of inhibiting cell growth, cell apoptosis, affecting protein expression, etc., and achieve the effect of strong specificity, efficient target identification, and rapid screening.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Target Screening Results
[0036] The applicant screened a large number of targets in the GS gene sequence of CHO cells, some of which are listed in Table 1.
[0037] Table 1. Preliminary optimized target sequence information for GS gene
[0038]
Embodiment 2
[0039] Construction and cell transfection of embodiment 2 short peptide coding sequence
[0040] According to the target information obtained in Example 1, carry out short peptide construction, and its specific construction process is as follows:
[0041] 1) Construction of plasmids containing TALE modules: commercially purchased TALE modules, a total of four single module plasmids (plasmid modules are: pMD-TALE-A (a TALE module plasmid that recognizes base A), pMD-TALE- T (TALE module plasmid that recognizes base T) pMD-TALE-C (TALE module plasmid that recognizes base C) pMD-TALE-G (TALE module plasmid that recognizes base G)), 16 double module plasmids (by Two single modules were assembled by enzyme-cut ligation). Each single module contains a repeat variable double residue (RVD) unit domain that specifically recognizes a single nucleotide base, specifically: NI recognizes A, NG recognizes T, HD recognizes C, NN recognizes G, and module domains Set SpeI site on one side, a...
Embodiment 3
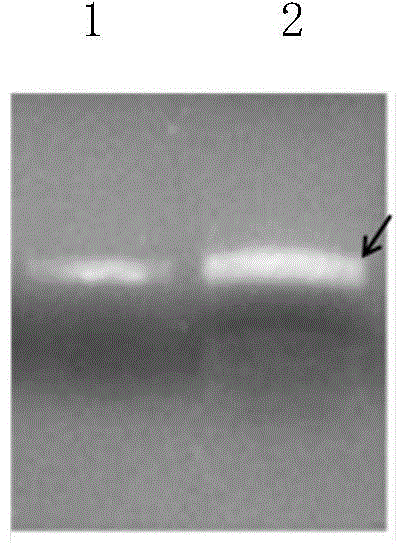
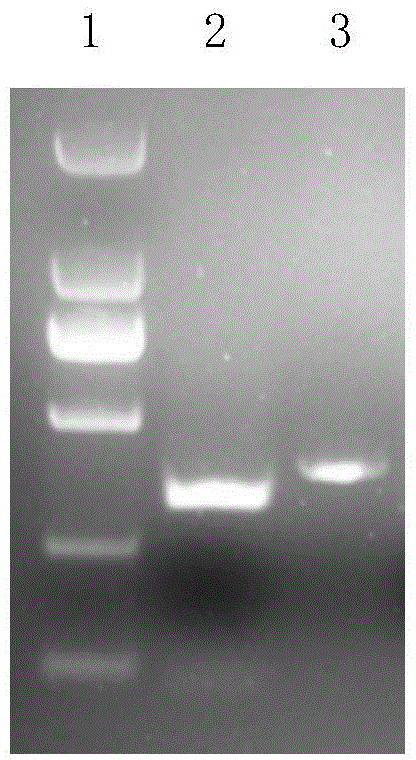
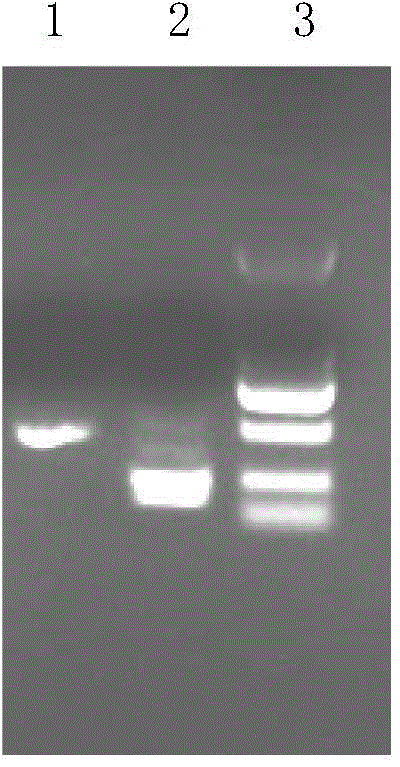
[0088] Identification of embodiment 3TALEN plasmid activity
[0089] 1) Centrifuge the transfected CHO-S cells to harvest the cells, perform DNA extraction according to the Qiagen Genome Extraction Kit, and set aside.
[0090]2) Using the extracted genomic DNA as a template for PCR reaction. Among them, target 1 corresponds to the CHO-S cell genomic DNA primer pair after vector transfection is primer 1 (5'-CCATGAGATTTCATAGCTG-3'), primer 2 (5'-GCAGATGGTCAGAGAATAAC-3'), and the peptides obtained from targets 2 and 3 After the sequence vector was transfected, the genomic DNA primer pair was primer 3 (5'-GACCTTGTCATTTTTCTGGCT-3') and primer 4 (5'-TGGAATTCCCACTAGAAAGAAC-3'). Target 1 corresponds to the PCR reaction conditions of CHO-S cell genomic DNA after vector transfection:
[0091] PCR reaction system:
[0092] 10× Taq enzyme buffer 2 μl
[0093] Primer 1 (10 μM) 0.8 μl
[0094] Primer 2 (10 μM) 0.8 μl
[0095] dNTP1.6μl
[0096] DNA template 50-100ng
[0097] r-Taq e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com