Specific primer pair for detecting shrunken4 gene of corn and application of primer pair
A technology of specificity and primer set, which is applied in the direction of recombinant DNA technology, microbial measurement/testing, biochemical equipment and methods, etc., can solve the problem of not obtaining co-dominant DNA molecular markers, etc., and achieve high accuracy and simple operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Acquisition of two co-dominant molecular markers
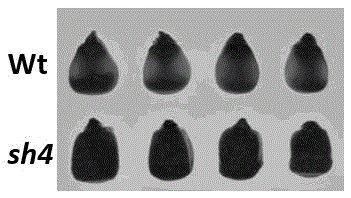
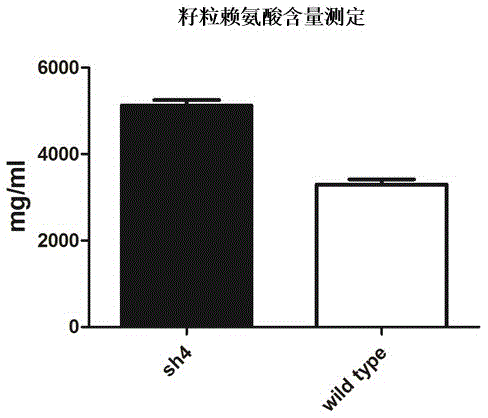
[0029] Since the sh4 mutant has a 55.43% increase in lysine (Lys) content compared to its corresponding wild type, see figure 2 .
[0030] The present invention knows that the sh4 gene is positioned on the long arm of the No. 5 chromosome of maize (Bin5.04-Bin5.05) through classical genetic mapping. Through the F2 population constructed for the homozygous mutant and homozygous wild-type plants near the sh4 site, the genomic DNA of each individual plant of the homozygous mutant, wild-type and F2 population was extracted. On the basis of previous rough mapping, the polymorphic SSR type molecular markers (umc2164 and umc1524) were obtained by screening the existing SSR molecular markers, and the sh4 gene was located between the molecular markers umc2164 and umc1524. However, the linkage between these two markers and sh4 is not very close, and there are still some problems in practical use. Therefore, based o...
Embodiment 2
[0038] Example 2: Determination of the linkage relationship between molecular markers InDel575, InDel592 and sh4
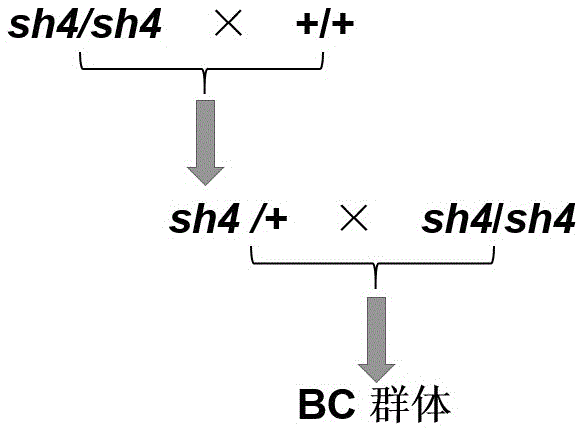
[0039] The DNA of 4 grains and mutant grains in the F1 population obtained by crossing sh4 / sh4 with the B73 background were extracted for the analysis of the linkage relationship between molecular markers InDel575 and InDel592 and sh4, and the genotype of the wild-type phenotype was sh4 / +, the genotype of sh4 homozygous mutants is sh4 / sh4. These 8 samples were used to analyze the genetic linkage relationship between the molecular markers InDel575 and InDel592 and the sh4 gene (see attached Figure 4 with 5 ). The results of the analysis showed that the two markers were linked to the sh4 gene, and the large population analysis showed that the exchange rate of the marker InDel575 was 3.51×10 in the segregated population of the B73 background. -4 , the exchange rate of labeled InDel592 is 8.77×10 -5 . For the physical location relationship between the molecular...
Embodiment 3
[0040] Example 3: Polymorphism identification of molecular markers among different parents
[0041] Genomic DNA of sh4 and six widely used inbred lines Chang7-2, W22, W64A, B73, Zheng58, and BSSS53 were extracted, and the polymorphism among different varieties was analyzed by PCR reaction. The results show that sh4 can be distinguished from all other inbred lines ( Figure 7 with Figure 8 ). This result proves that the molecular markers InDel575 and InDel592 can accurately detect the existence of the sh4 gene in any breeding background population, and the misselection rate during assisted selection is 3.08×10 -8 , which can satisfy the selection requirement.
[0042] Shanghai University
[0043] Specific primer set for detecting maize shrunken4 gene and its application
[0044] 4
[0045] 1
[0046] 18
[0047] ssDNA
[0048] Artificial sequence
[0049] 1
[0050] GGAAAGCGAAGCAGGTAA18
[0051] 2
[0052] 20
[0053] ssDNA
[0054] Artificial sequence...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com