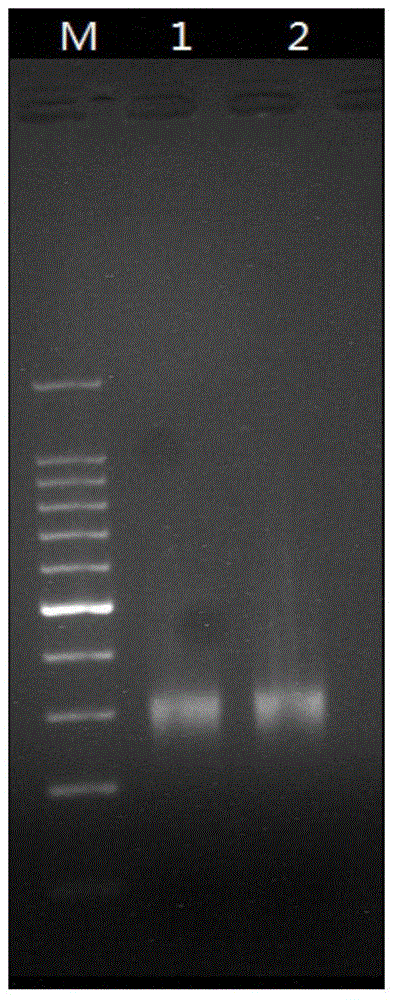
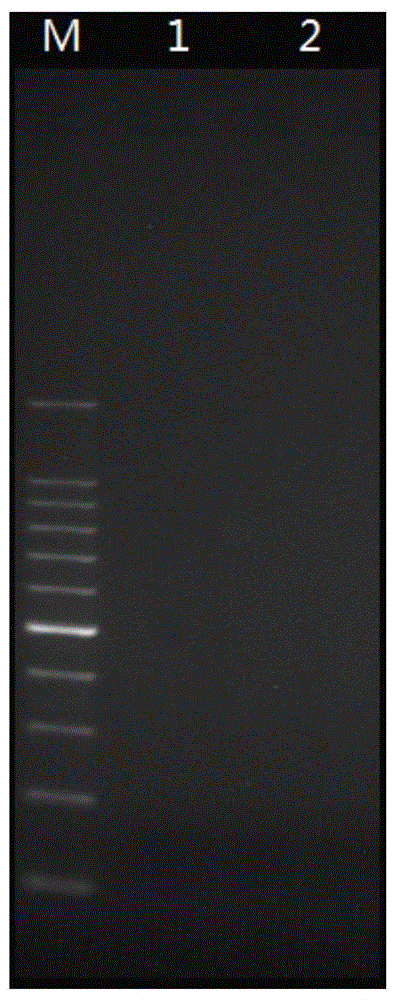
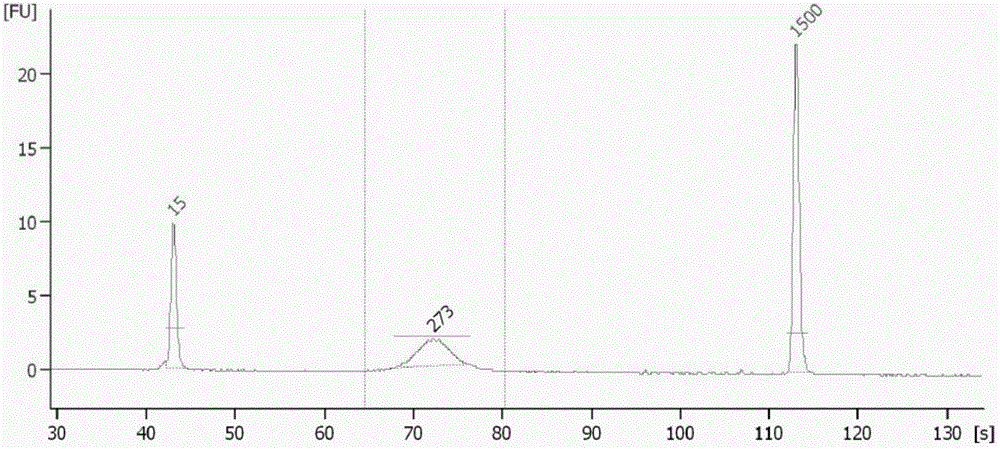
PCR amplification system and library construction method for whole genome bisulfate sequencing
A bisulfite, whole genome technology, applied in the field of molecular biology, can solve the problems of unreachable detection concentration, few PCR products, low DNA concentration, etc., to reduce workload, improve work efficiency, and reduce consumption. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Reagents and kits used in this embodiment:
[0035] (1) Agilent: high-fidelity CX hot start DNA polymerase (PfuTurbocxHotstartDNAPolymerase)
[0036] (2) Germany Kaijie (QIAGEN): bisulfite treatment kit (EpiTectBisulfiteKit)
[0037] (3) IllumineDNA Library Construction Kit (TruSeqDNASamplePreparationLowThroughput(LT)Kit)
[0038] 1. Quantification of DNA input amount
[0039] Note: The starting amount of DNA used in this experiment is 5 μg.
[0040] 2. DNA Fragmentation
[0041] (1) Turn on the ultrasonic interrupter in advance and cool down to below 6°C;
[0042] (2) DNA sample (tiger frog virus (TFV) DNA) was diluted to 130 μl with 1×TE, mixed with a gun, transferred all the solution to a clean and dry atomizing cup, and interrupted to 200 bp.
[0043] 3. Purification of DNA Fragments
[0044] (1) Add 3 times the volume of PCR-A solution to the fragmented DNA, mix well and transfer to a purification column up to 700 μl each time, centrifuge at 10,000 rpm for 1 ...
Embodiment 2 comparative test example
[0160] A methylation library for whole-genome bisulfite sequencing was prepared by a conventional method, and the rest of the steps were the same as in Example 1, except that in operation step 10, that is, during PCR amplification, the following conventional PCR amplification system was used:
[0161] Aliquot 12 μl of salt-treated and purified DNA into 3 PCR tubes, and the reaction system in each tube is:
[0162] DNA4μl
[0163] UltraPureWater33.75μl
[0164] PfuTurboCxReactionBuffer5μl
[0165] 10mMdNTPMix 1.25μl
[0166] PCR Primer Cocktail 5 μl
[0167] PfuTurboCxHotstartDNAPolymerase 1 μl
[0168] Total Volume50μl
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com