Detection method of microRNA (micro Ribonucleic Acid)
A detection method and polyuracil technology, applied in the field of microRNA detection, can solve the problem that the detection technology has not yet been reported, and achieve the effect of low background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] (1) Double polymerase extension of uracil sequence: mix microRNA and template DNA (pDNA), anneal and hybridize at 75°C for 15 minutes, then slowly cool to room temperature, add polymerase-containing Klenow fragment (KFexo - ), deoxyuridine triphosphate and terminal deoxynucleotidyl transferase (buffer solution, reacted in a water bath at 35°C for 4 hours to prepare a polyuracil base sequence.
[0020] (2) Synthesis of fluorescent copper nanoclusters: Add ascorbate to the polyuracil base sequence solution in step (1), then add CuCl 2 , and react at room temperature for 10 minutes to prepare red fluorescent copper nanoclusters with polyuracil base sequence as template.
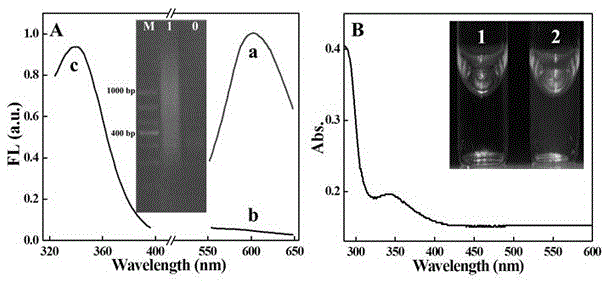
[0021] Depend on figure 1 A shows that when microRNA-21 exists, the CuNCs synthesized in situ using the polyuracil base sequence formed by the extension of uracil by double polymerases as a template has a strong fluorescence emission peak at 600nm (curve a), and the optimal excitation wavelength is 340n...
Embodiment 2
[0027] According to the research process of Example 1, the microRNA detection method was determined.
[0028] (1) Double polymerase extension of uracil sequence: Mix microRNA with known content and template DNA (pDNA), anneal and hybridize at 75°C for 15 minutes, then slowly cool to room temperature, add polymerase-containing Klenow fragment (KFexo - ), a buffer solution of deoxyuridine triphosphate and terminal deoxynucleotidyl transferase (TdTase), and react in a water bath at 35°C for 4 hours to prepare a polyuracil base sequence;
[0029] (2) Synthesis of fluorescent copper nanoclusters: add ascorbate to the polyuracil base sequence solution in step (1), and then add CuCl 2 , reacting at room temperature for 10 minutes, making red fluorescent copper nanoclusters with polyuracil base sequence as template, and obtaining a solution containing red fluorescent copper nanoclusters;
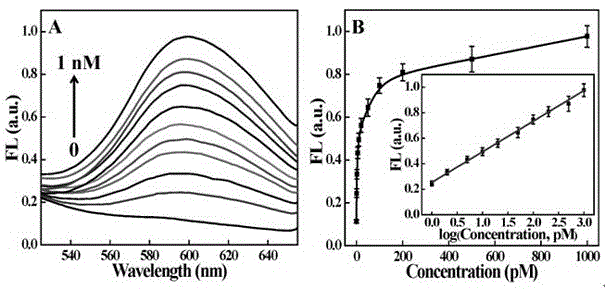
[0030] (3) Use a fluorescence spectrophotometer to calibrate the standard relationship curve be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com