OsZFP gene and application of OsZFP gene protein to rice lateral root growth and development regulation and control
A growth and genetic technology, applied in the field of molecular biology, can solve the problems of yeast toxicity, unsuitability for yeast two-hybrid screening system, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] 1. PCR amplification of OsCYP2 gene
[0047] Design PCR amplification primers BD-OsCYP2-F and BD-OsCYP2-R according to the multiple cloning sites (EcoRI and SalI) of the yeast expression vector pGBKT7 and the OsCYP2 gene sequence (519bp), as follows (the underline is the restriction site):
[0048] BD-OsCYP2-F:CGG GAATTC TCTGTGAAATTCGCAAAACCC (SEQ ID NO. 1)
[0049] BD-OsCYP2-R:ACGC GTC GAC ACACCACCACCACCTCCTCCT (SEQ ID NO. 2)
[0050] The OsCYP2 gene was amplified by PCR using the first-strand cDNA of the rice variety 'Aichi Asahi' as a template, and the reaction procedure was as follows:
[0051]
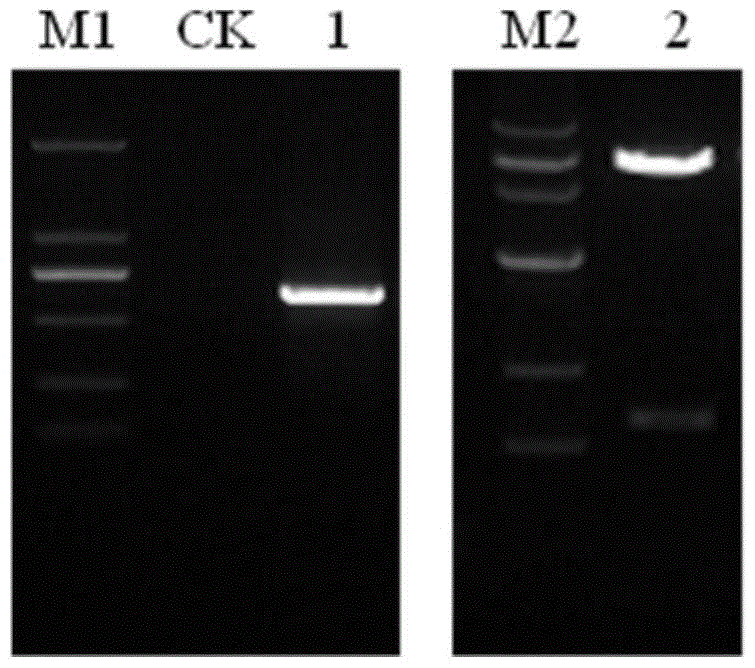
[0052] PCR amplification products were detected by agarose gel electrophoresis and photographed in BIO-RAD gel imager (such as figure 1 shown). The target fragment and the yeast expression vector pGBKT7 were subjected to double digestion (EcoRI and SalI), the target fragment and the vector backbone were recovered, and T4 DNA ligase was used to ligate overnight at ...
Embodiment 2
[0102] 1. Construction of OsZFP gene interference expression vector
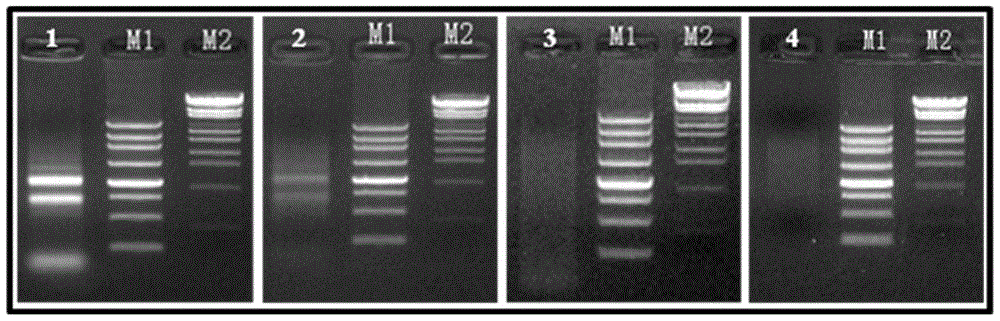
[0103] According to the interference expression vector pCAMBIA1300-RNAi multiple cloning site and the OsZFP gene target site sequence (264bp), the forward insert PCR amplification primers RNAi1-F and RNAi1-R, and the reverse insert PCR amplification primers RNAi3-F and RNAi3-R, the schematic diagram of the construction of the interference expression vector is as follows: Figure 7 As shown, the relevant primer sequences are as follows (the underline is the restriction site):
[0104] RNAi1-F:CGC GGATCC TGTAAAATATGTGGAGAATCAGGTC (SEQ ID NO. 7)
[0105] RNAi1-R:GG ACTAGT CTCCTGAATGATTTTATGAACAGCA (SEQ ID NO. 8)
[0106] RNAi3-F:C GAGCTC TGTAAAATATGTGGAGAATCAGGTC (SEQ ID NO.9)
[0107] RNAi3-R:CGG GGTACC CTCCTGAATGATTTTATGAACAGCA (SEQ ID NO. 10).
[0108] OsZFP gene target sequence PCR amplification reaction procedure is as follows:
[0109]
[0110]
[0111]PCR amplification products were de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com