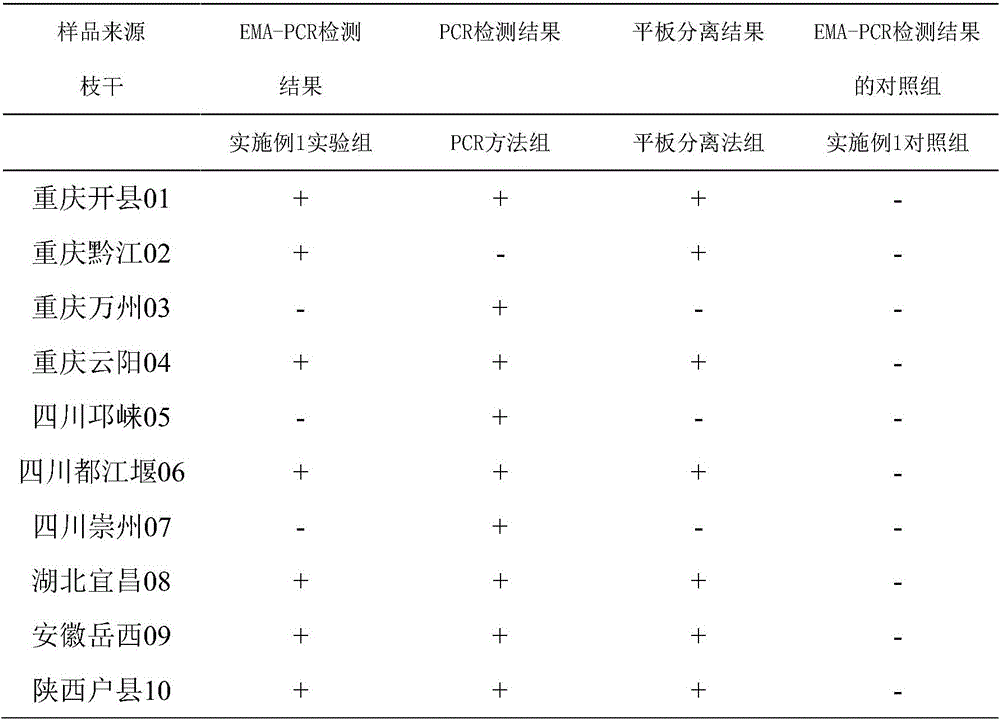
Living pseudomonas syringae pv.actinidiae detection method
A technology for canker bacteria and kiwifruit, which is applied in the field of detecting live bacteria of kiwifruit canker bacteria, can solve the problems of difficult operation and promotion, difficult detection of target bacteria, false positive detection, etc., so as to reduce the steps of cell lysis and DNA extraction, and save detection time. and cost, reducing false positives and false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Reagents: EMA (Invitrogen), Taq polymerase (TaKaRa), and conventional PCR reagents were purchased from Shanghai Biyuntian Biotechnology Co., Ltd.
[0025] A method for detecting live bacteria of kiwifruit canker bacterium, which is operated according to the following steps:
[0026] (1) Clean the branches of the kiwi fruit to be tested and cut them into small pieces of 4 mm × 4 mm, wash them with sterile water for 3 times, then disinfect them with 1% sodium hypochlorite and 70% ethanol for 1 min, and rinse them with sterile water for 3 times;
[0027] (2) After crushing the sample under sterile conditions, add 0.8 mL of sterile distilled water to soak for 2 hours, take 0.4 mL of the upper soaking solution and inoculate it into 10 mL of LB liquid medium, and incubate on a shaking table at 25°C and 250 r / min for 8 hours;
[0028] (3) Take the culture solution after proliferation culture and add EMA at a final concentration of 3 μg / mL in the dark; treat the culture solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com