Primer, kit and method for detecting CHO cell DNA residues
A kit and cell technology, applied in the field of quantitative detection of DNA content in CHO cells, can solve the problems of comparable specificity, not widely used, and low success rate, and achieve the effect of high specificity and good sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
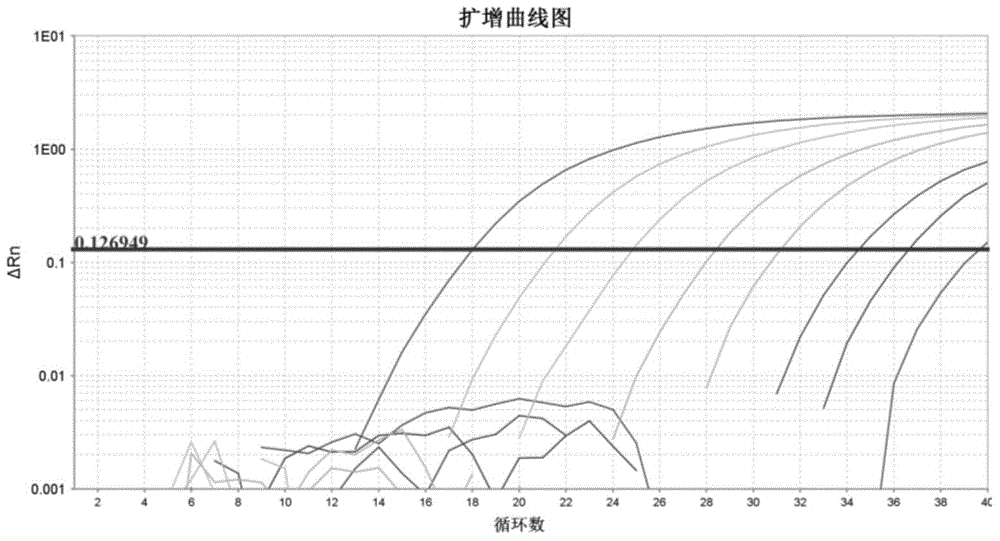
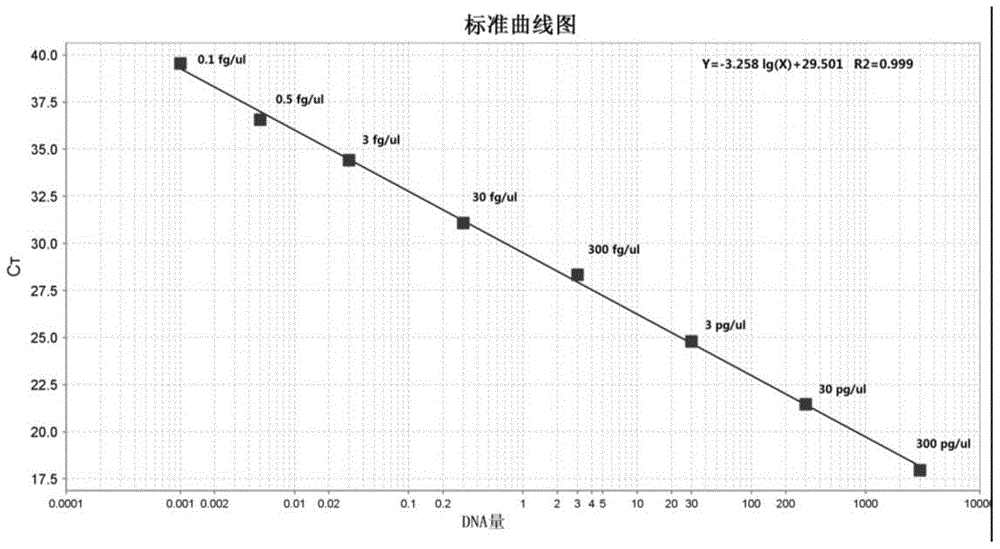
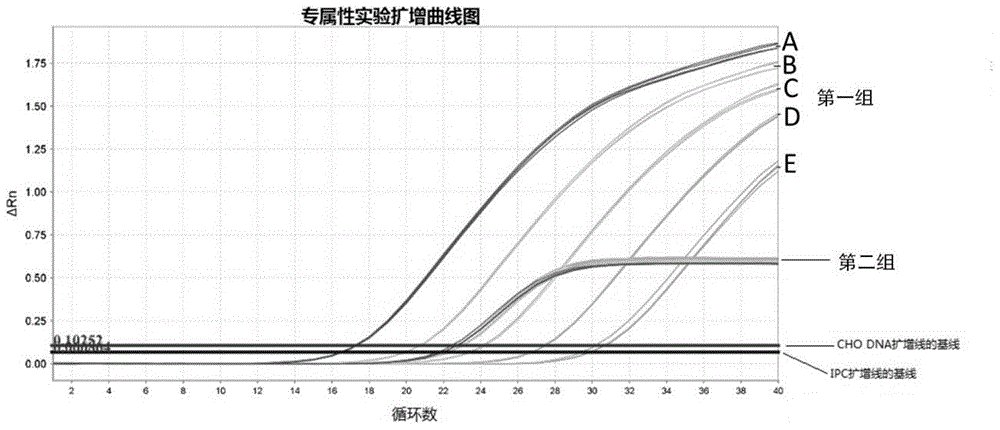
[0050] Example 1: Methodological Confirmation
[0051] 1.1 Materials and reagents: 2×Q-PCR master mix is a commercially available product, the ViiA 7 fluorescent quantitative PCR instrument is a product of Life Technologies, the DNA dilution is prepared by our laboratory using molecular biology grade reagents, and the genomic DNA extraction kit is a product of Life Technologies ; The IPC template in the IPC reagent is the PGEM-T Easy Vector series product of Promage Company, and the primers and corresponding probes involved in the experiment are synthesized by Life Technologies Company.
[0052] The primer sequences and probe sequences used for methodological confirmation are as follows:
[0053] Forward primer sequence: 5'-CTACCAGAGGTCCTGAGTTCAATT-3' (SEQ ID NO: 1)
[0054] Reverse primer sequence: 5'-GGGCAC CAGGTCTCATAACG-3' (SEQ ID NO:2)
[0055] The nucleotide sequence of the Taqman probe is:
[0056] Taqman probe: 5'-CCAGCAACCA CATGGTGGCT CAC-3' (SEQ ID NO: 3)
[...
Embodiment 2
[0093] The preparation of embodiment 2 kit
[0094] Prepare a kit comprising the following components:
[0095] DNA diluent (5mL / tube) 1 tube, Q-PCR master mix (1.5mL / tube) 1 tube, IPC reagent (250μl / tube) 1 tube, CHO cell DNA standard (40μl / tube) 1 tube, forward Primer (100 μl / tube) 1 tube, reverse primer (100 μl / tube) 1 tube, Taqman probe (50 μl / tube) 1 tube, pure water (1 mL / tube) 1 tube.
[0096] CHO cell DNA standard, the concentration is 3.0×10 7 fg / μl.
[0097] IPC reagent: Each 2.5 μl IPC reagent contains 1 μl 1fg / μl IPC template, 0.5 μl 10 μM IPC-F, 0.5 μl 10 μM IPC-R, 0.5 μl 5 μM IPC-Probe.
[0098] The forward primer, reverse primer, and Taqman probe are all 10 μM.
[0099] DNA diluent: use deionized water as solvent, Tris-HCl concentration is 10mM / L, EDTA concentration is 0.5mM / L, adjust pH to 8.0 with NaOH solution.
[0100] 2×Q-PCR master mix: Q-PCR master mix (2×).
[0101]
[0102]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com