Enzymatic activity improved pyruvate carboxylase mutant N1078F and application thereof
A pyruvate carboxylase and mutant technology, which is applied in the fields of genetic engineering and fermentation engineering, can solve the problems of carbon flow loss, lack of carboxylic acid synthesis pathway, etc., and achieves the effects of improving yield, good industrial application value and prospect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
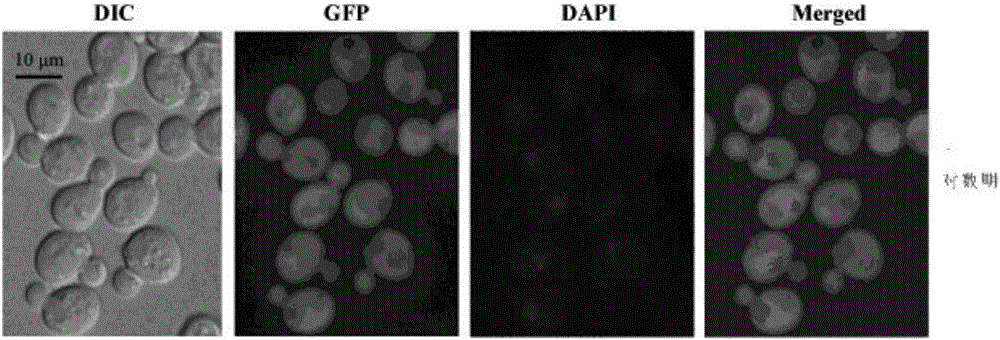
[0027] Example 1: Localization of RoPYC in Saccharomyces cerevisiae
[0028] 1. TEF1 promoter and RoPYC gene ORF were cloned into pGFP33 vector
[0029] According to the principle of the homologous recombination kit, design primers with more than 15 bases homologous to the vector at both ends, as shown in Table 1, the lowercase letters are homologous arms, and the uppercase letters are PCR primers.
[0030] Table 1 Primers for amplifying RoPYC gene
[0031]
[0032] Using pY15TEF1-RoPYC as a template (Xu et al. Fumaric acid production in Saccharomyces cerevisiaeby in silico aided metabolic engineering, 2012), RoPYC-F, RoPYC-R (sequences are shown in SEQ ID NO.3, SEQ ID NO.4 respectively ) for primers to amplify the ORF frame of TEF1p and RoPYC (the obtained PCR product contains the sequence of the amino acid sequence SEQ ID NO.1), and the amplified product is connected to the pGFP33 vector (a low-copy expression with green fluorescent protein GFP carrier, using a fluoresc...
Embodiment 2
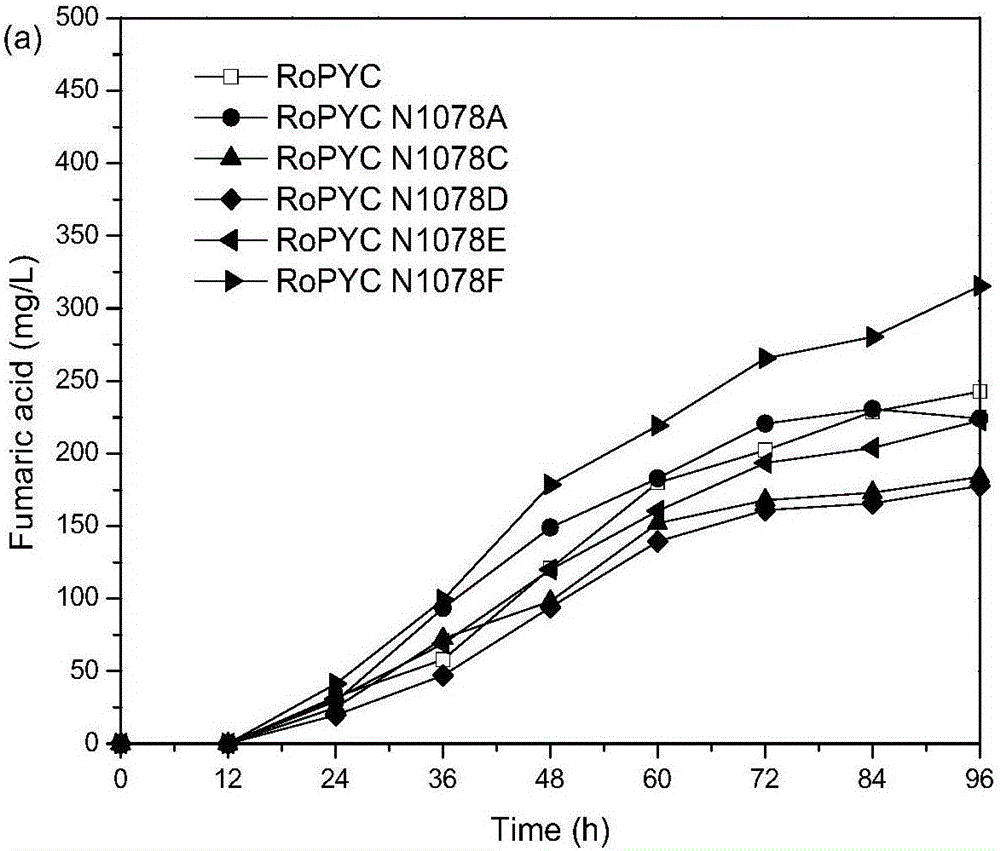
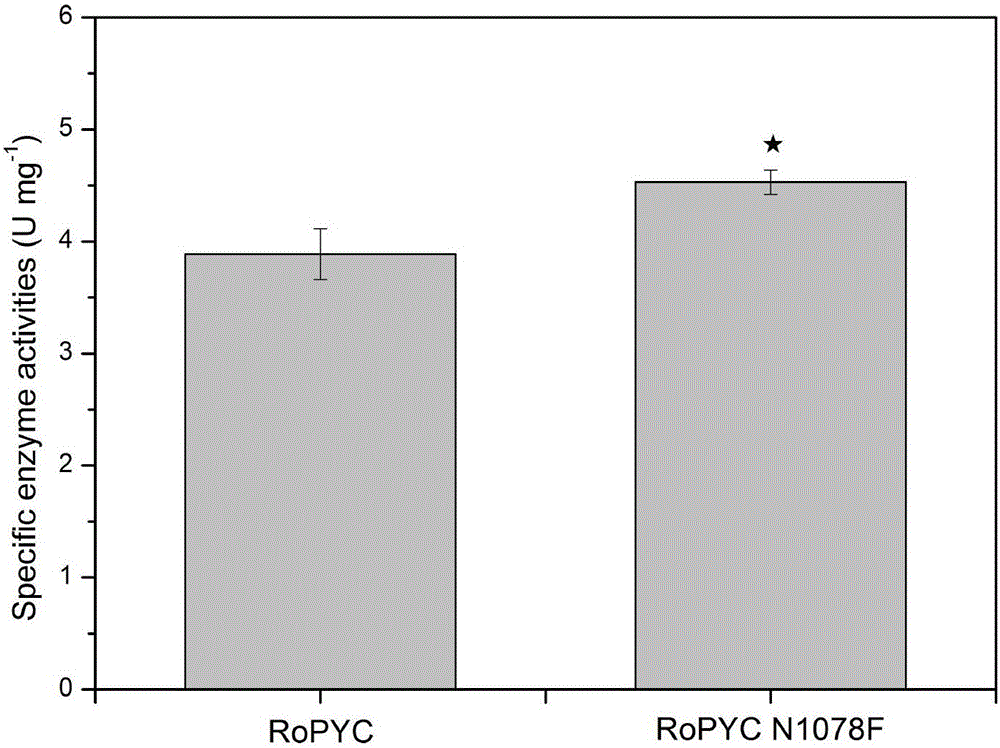
[0037] Example 2: RoPYC site-directed mutation, expression and production of fumaric acid
[0038] Site-directed mutagenesis was performed by PCR method, and the pY15TEF1-RoPYC plasmid was used as a template, F and R containing the mutation site were used as primers, and the high-fidelity enzyme PrimeSTAR GXL of Takara Company was used to perform PCR to amplify the entire plasmid. The enzyme digestion system contains 1 μL of PCR product and 1 μL of Dpn I enzyme, with a total volume of 20 μL, digested overnight at 37°C. Fragment purification of digested products. Take 5 μL of the purified product and transform it into 30 μL of competent cells Trans1-T1, smear the LA plate, inoculate the grown transformant in the LA medium, extract the plasmid and send it to Shanghai Sangon for sequencing.
[0039] Among them, the primers F(Asn) and R(Asn) (sequences shown in SEQ ID NO.5 and SEQ ID NO.6 respectively) used for N1078F mutation are shown in Table 2.
[0040] Table 2 Primers for s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com