SNP (single nucleotide polymorphism) marker related to hevea brasiliensis stem girth and application thereof
A rubber tree and labeling technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems that cannot meet the needs of molecular breeding of rubber trees, achieve the effect of convenient operation and promote the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1: Obtaining of SNP markers related to rubber tree stem girth
[0031] 1.1 Acquisition of rubber tree germplasm materials
[0032] The population used is the rubber tree 1981'IRRDB germplasm, which was planted in the National Rubber Tree Germplasm Resource Garden of the Rubber Research Institute of the Chinese Academy of Tropical Agricultural Sciences. Mato Grosso and Rondonia three states. In June 2014, 34 germplasm young leaves were collected and frozen in liquid nitrogen and brought back to the laboratory for future use.
[0033] 1.2 DNA extraction of rubber tree germplasm material
[0034] Take the frozen rubber tree leaves, and use the CTAB method to extract genomic DNA: (1) Weigh 1g of rubber tree leaves, freeze them in liquid nitrogen, grind them into fine powder, and transfer them to a 50ml centrifuge tube. (2) Add 10ml of 65°C preheated 2×CTAB extraction buffer (2% β-mercaptoethanol has been added in advance), gently rotate the centrifuge tube t...
Embodiment 2
[0037] Example 2: Sequencing verification and application of SNP markers related to rubber tree stem circumference
[0038] 2.1 Amplification of nucleotide fragments containing SNP sites
[0039] Using the genomic DNA of each rubber tree to be tested obtained by the aforementioned extraction as a template, use forward primer F: 5'-GAA GCA CTA AGA CGG TCC ATA G-3' (SEQ ID NO: 2) and reverse primer R: 5' - GGA GAC AGA TTA CAA TCT GAC AGC-3' (SEQ ID NO: 3), amplifying the nucleotide fragment where the SNP to be detected is located. Among them, the PCR reaction system is 25 μl: 1 μl of 100-200ng / μl template DNA, 1 μl of 10 μM primers F and R, 2.5 μl of 10×PCR reaction buffer, 2.0 μl of 2.5mM dNTP, 0.2 μl of 5U / μl Tap DNA polymerase, Water balance; PCR reaction conditions: 95°C pre-denaturation for 5 minutes, 30 cycles at 94°C for 30s, 60°C for 30s, 72°C for 1min, and extension at 72°C for 5min.
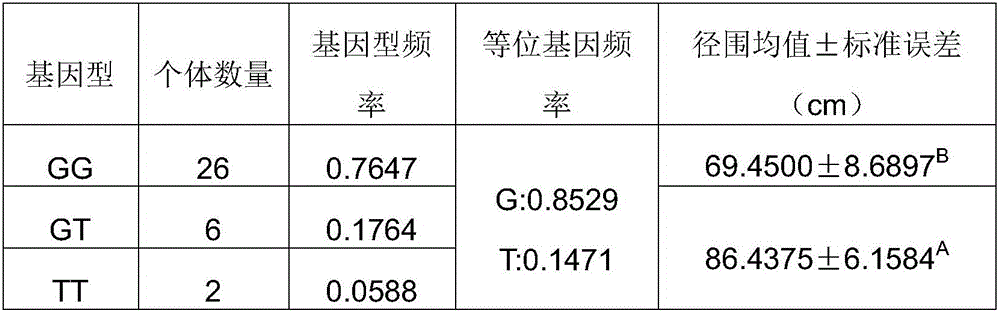
[0040] 2.2 Sequencing to identify the genotype of the SNP site
[0041] The PCR pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com