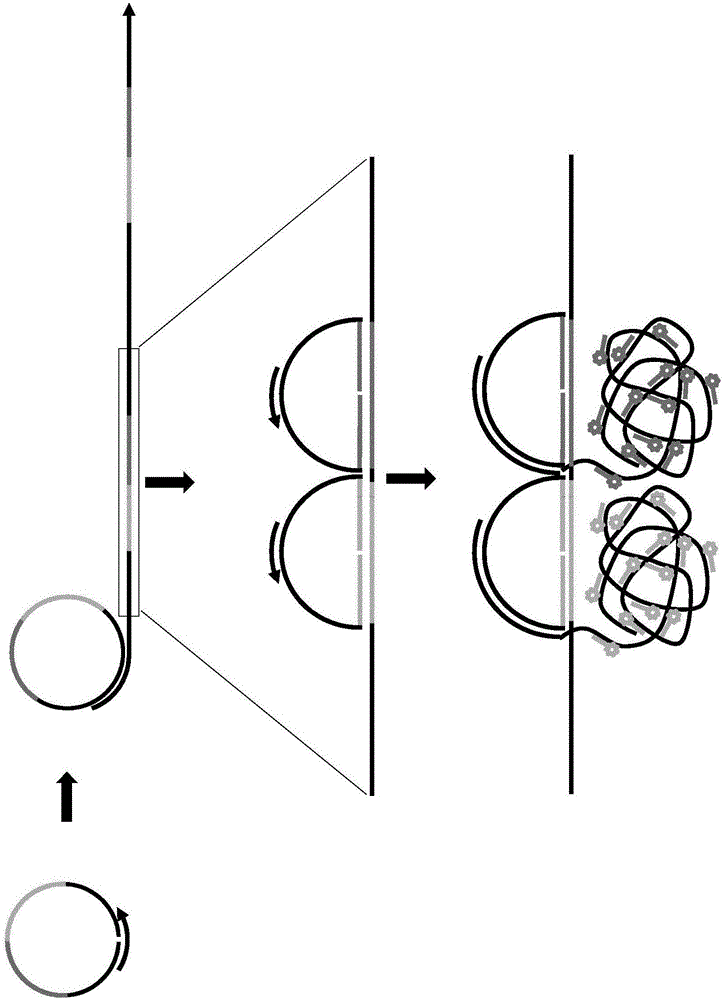
Localised Rca-Based Amplification Method Using A Padlock-Probe
A local, product technology, applied in the conceptual field of local highly linear amplification, to achieve the effects of enhanced signal amplification, enhanced sensitivity, and fast signal generation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
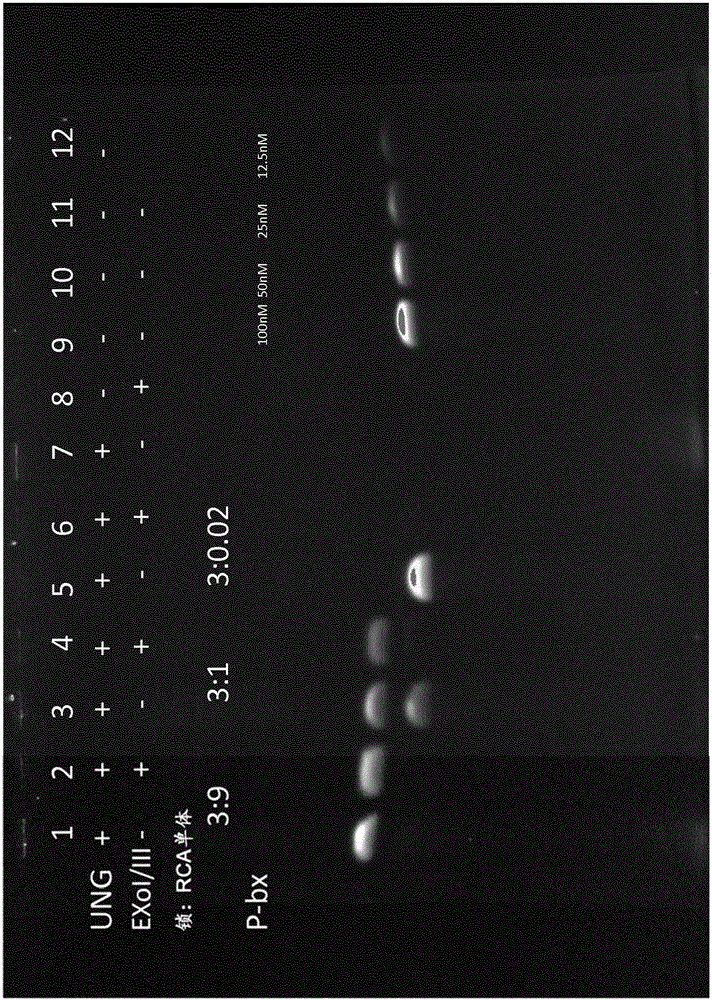
[0197] This example investigates linkage and efficiency and effects of different lock:RCA monomer ratios.
[0198] The approximate experimental setup is as follows: The first RCA is generated by using a set of dNTPs containing dUTP. Different lock concentrations and linker components are then added. After ligation, the reaction mixture was treated with UNG or UNG+Exonuclease I / Ill to degrade the first RCA product or the first RCA product and any unligated linear locks. Samples were then analyzed on 6% TBE-urea gels.
[0199] Several different amounts of pre-generated uracil-containing 1st RCA products were added to 50 μl 2nd lock ligation mix (which consisted of 1X buffer 4 (New England Biolabs), 0.2 mM ATP, 0.02 U T4 DNA ligase (Thermo -Scientific) and 0.02mg / ml BSA (New England Biolabs) and 100nM p-bx locked oligonucleotide (SEQ ID No.11) composition) to obtain the first 3:9, 3:1 or 3:0.02 2RCA lock: the first RCA monomer. Reactions were incubated at 37°C for 30 minutes....
Embodiment 2
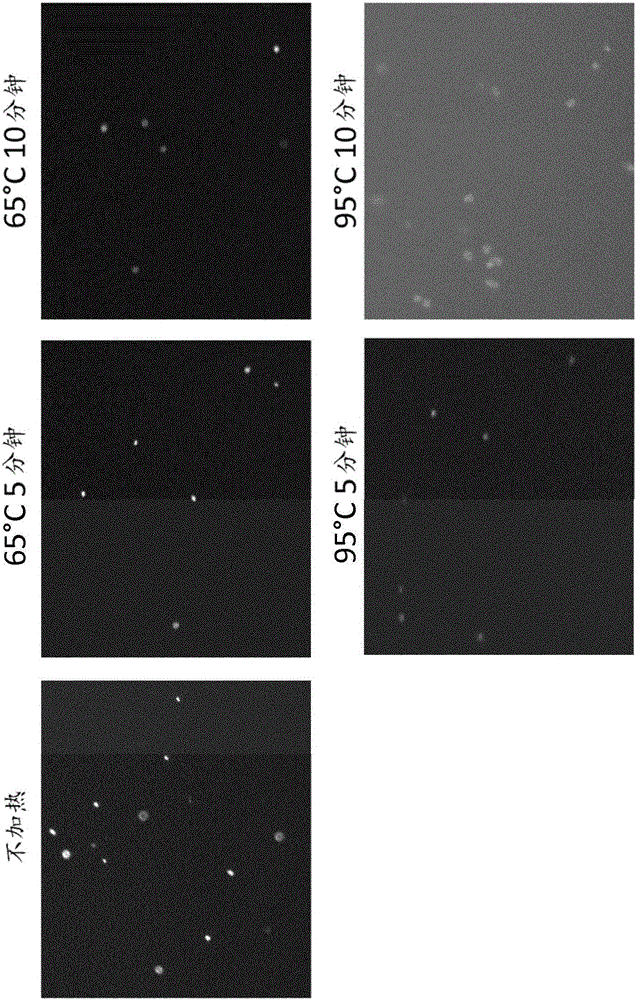
[0201] stability test
[0202] This example was performed to test the stability of the lock sRCA at high temperature ex 65°C and 95°C. 1 μl of the first RCA product was added to 50 μl of the second lock ligation mix (made with 1X buffer 4 (New England Biolabs), 0.2 mM ATP, 0.02 U T4 DNA ligase (Thermo-Scientific) and 0.02 mg / ml BSA (New EnglandBiolabs) and 100nM p-bx locked oligonucleotide (SEQ ID NO.11) composition). Reactions were incubated at 37°C for 30 minutes. Then 1 mM dNTP, 300 nM sRCA-x-primer oligonucleotide and 200 nM sRCA-d-1b-Cy3 oligonucleotide and 0.1 U phi29 polymerase (Thermo-Scientific) were added to the reaction mixture, followed by incubation at 37° C. Incubate for 100 minutes. The reaction mixture was then divided into 5 parts, and each part was subjected to different heat treatment conditions. 10 [mu]l of each heated sample was then placed on a poly-L-lysine slide and visualized with an EPI fluorescence microscope (Carl Zeiss). The results are show...
Embodiment 3
[0204] Experiments compared the results of different rounds of RCA using padlock probes and showed the signal amplification achievable with padlocked sRCA; stronger signals were seen for 2 (2RCA) or 3 (3RCA) rounds of RCA.
[0205]Streptavidin-coated glass slides (Surmodics) were incubated with 50 μl of 1 pM primary lock ligation mixture for 30 minutes at 37° C., followed by 2 washes with 50 μl of 1 XPBS + 0.05% Tween 20. 50 μl of RCA mix (consisting of 1X Buffer 4 (New England Biolabs), 1 mM dNTPs, 0.1 U phi29 polymerase (Thermo-Scientific), 0.02 mg / ml BSA (New England Biolabs)) was then applied to the slide, And incubated at 37° C. for 20 minutes, then washed twice with 50 μl of 1XPBS+0.05% Tween 20. Afterwards, 50 μl of ligation mix (made of 1X buffer 4 (New England Biolabs), 0.2 mM ATP, 0.02 U T4 DNA ligase (Thermo-Scientific) and 0.02 mg / ml BSA (New England Biolabs) and 100 nM p-bx Locked oligonucleotide (SEQ ID No. 11) was applied to the glass slide and incubated at 37°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com