Two black mustard genomes DNA sequence and application thereof
A genome and DNA molecule technology, applied to two black mustard genome DNA sequences and their application fields, can solve the problems of cumbersome methods, the identification is greatly influenced by subjective factors, and the repeatability is poor, so as to achieve the effect of easy identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
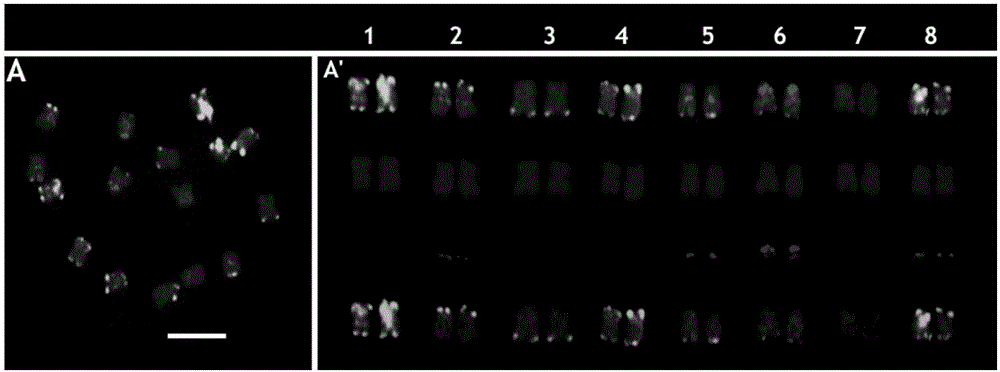
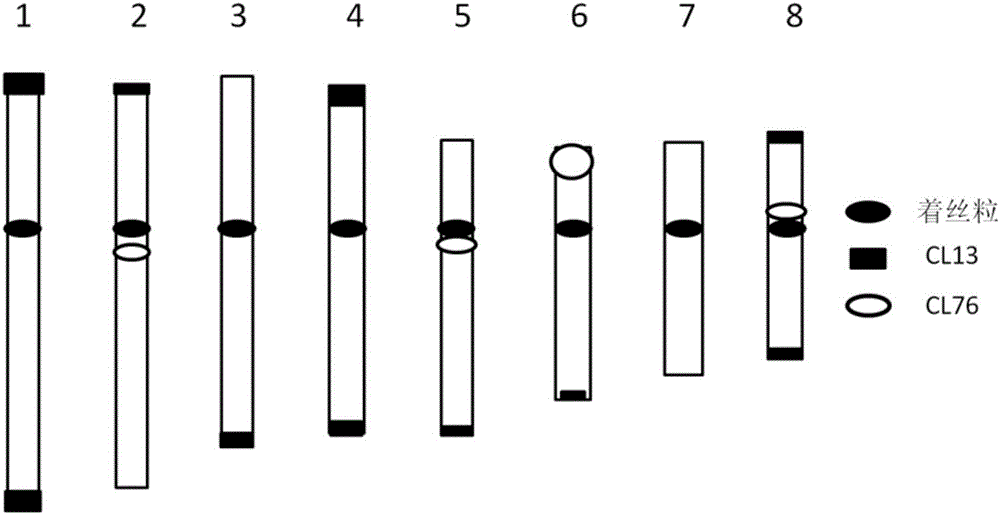
[0033] Example 1, the separation of DNA repeat sequence CL13 and CL76
[0034] Using black mustard 'G1 / 1' as a material, the base number of 0.6G obtained by random genome sequencing is equivalent to about 0.8 times of the black mustard genome (calculated based on the genome size of 630Mb, Johnston et al., 2005). Combining the methods of Novák et al. (2010) and Macas et al. (2010) for repeat sequence analysis, two tandem repeat sequences CL13 and CL76 with abundant genome content were found, and verified by PCR cloning.
[0035] The nucleotide sequence of CL13 is sequence 1 in the sequence listing.
[0036] The nucleotide sequence of CL76 is sequence 2 in the sequence listing.
Embodiment 2
[0037] Embodiment 2, the application of DNA repeat sequence CL13 and CL76 as detection probe
[0038] 1. Fluorescence in situ hybridization (FISH) test method using CL13 and CL76 as detection probes
[0039] 1. Chromosome production
[0040] 1) Put the plant seeds to be tested in a petri dish with moist filter paper, and take root at room temperature;
[0041] 2) When the root tip length is 0.5-1cm, cut off the root tip and put it into the d 2 h 2 O in a test tube, and put the test tube into an ice box filled with ice-water mixture and place it in the refrigerator for 22-24h for pretreatment;
[0042] 3) Put the pretreated root tip into fixative solution (absolute ethanol:glacial acetic acid=3:1 (v / v)) and fix it for 1-7 days;
[0043] Note: In order to obtain better results in in situ hybridization, the fixation time should not be too long. Of course, if the fixation time is less than two days, it is relatively difficult to make slices.
[0044] 4) Excise part of the roo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com