Construction method of microbial peptide mass fingerprint spectrum database based on time-of-flight mass spectrometry principle
A time-of-flight mass spectrometry and fingerprint technology, which is applied in biological testing, measuring devices, material inspection products, etc., can solve the problems that microorganisms cannot be identified, do not meet actual needs, and can only be identified to the genus level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
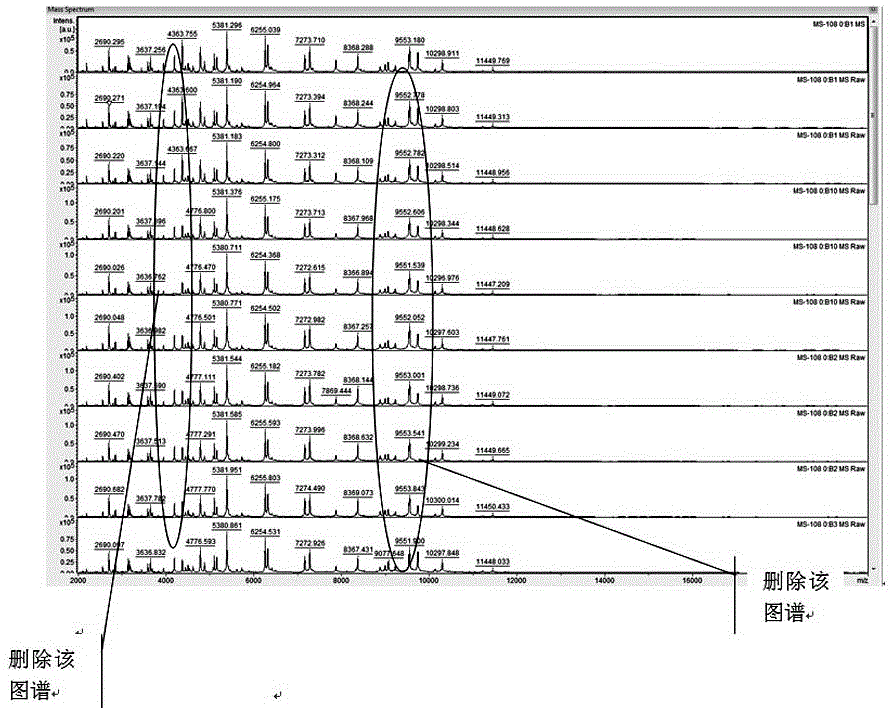
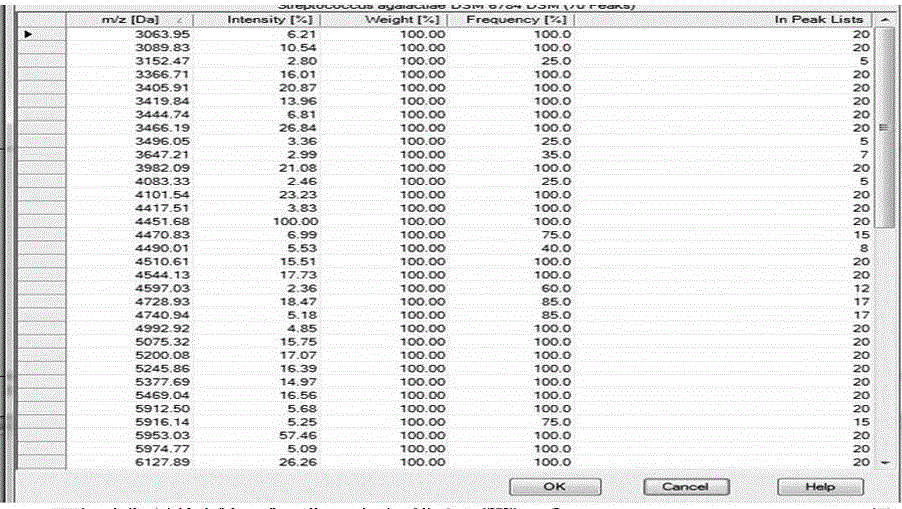
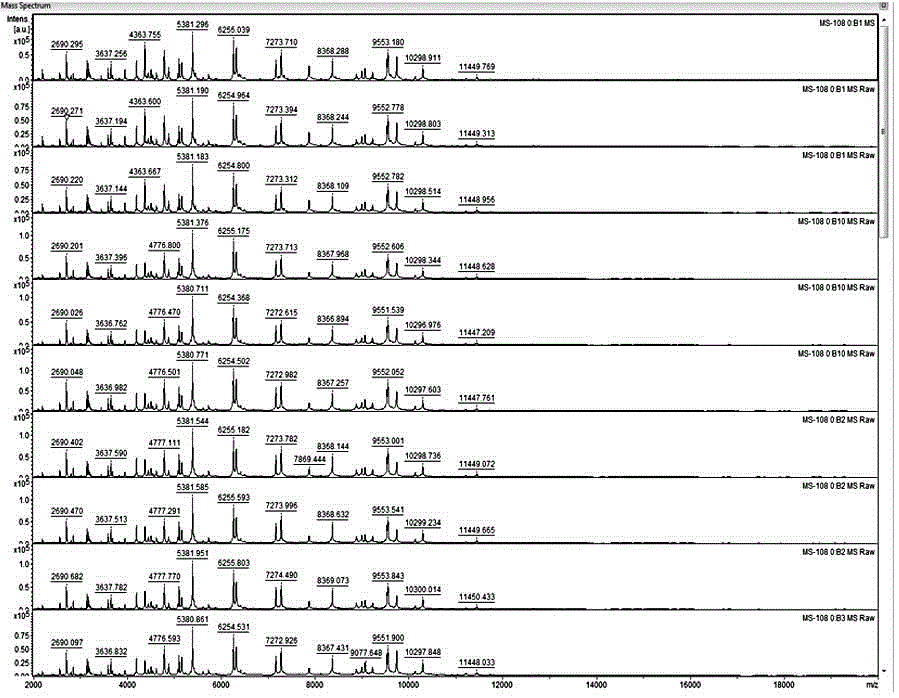
[0041] The construction method of the microbial peptide mass fingerprint library based on time-of-flight mass spectrometry principle of the present invention comprises the following detailed steps:
[0042] First step, standardization of strains
[0043] 1. Culture of strains
[0044]Firstly, the strains obtained from scientific research institutes, preservation centers, etc. were cultured on different media according to their physiological characteristics: for example, fungi were cultured on Sabouraud agar plates, bacteria were cultured on Columbia blood plates or ordinary blood plates at 37 degrees, respectively. Cultivate for 48h and 24h. Neisseria, Streptococcus, etc. need to be inoculated on chocolate agar plates, and cultured in a 5% carbon dioxide environment for 24h to obtain logarithmic phase strains. The principle is to strictly abide by the established conditions of various microorganisms. culture conditions, improve the identification accuracy of microorganisms; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com