Rapid mass spectrometry and spectrum identifying method for peptide fingerprint spectrum
A technology of fingerprint and mass spectrometry, applied in the direction of analyzing materials, material separation, measuring devices, etc., can solve the problems that hinder the clinical application of peptide spectrum analysis technology, lack of rapid analysis kits, etc., and achieve accurate, fast, simple and easy storage of detection methods , Improve the effect of detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] 1) Fix the P-type boron-doped crystalline silicon wafer in the electrolytic cell, add 55ml of ethanol with a volume ratio of 1:0.05-1:6 and 10%-40% hydrofluoric acid as the electrolyte, and The silicon wafer is used as the anode, and the platinum electrode is used as the cathode. Perform DC electrolytic etching and stripping. Set the current intensity to 0.2-2 1, and the etching time to 60-600 s. After vacuum drying for 10 minutes, porous silicon particles with a pore diameter of 1-15 nm, a thickness of 0.1-10 μm, and a diameter of 10-50 μm were obtained.
[0043] 2) Porous silicon particles modified by gold nanoparticles are prepared by electrochemical deposition, and the freshly prepared porous silicon chip is placed in a Teflon tank, and electrolyzed with chloroauric acid (0.01-0.1% w / v) solution. liquid for electrochemical deposition. After the completion, the porous silicon layer is peeled off, and the surface-modified gold porous silicon particles are obtained a...
Embodiment 2
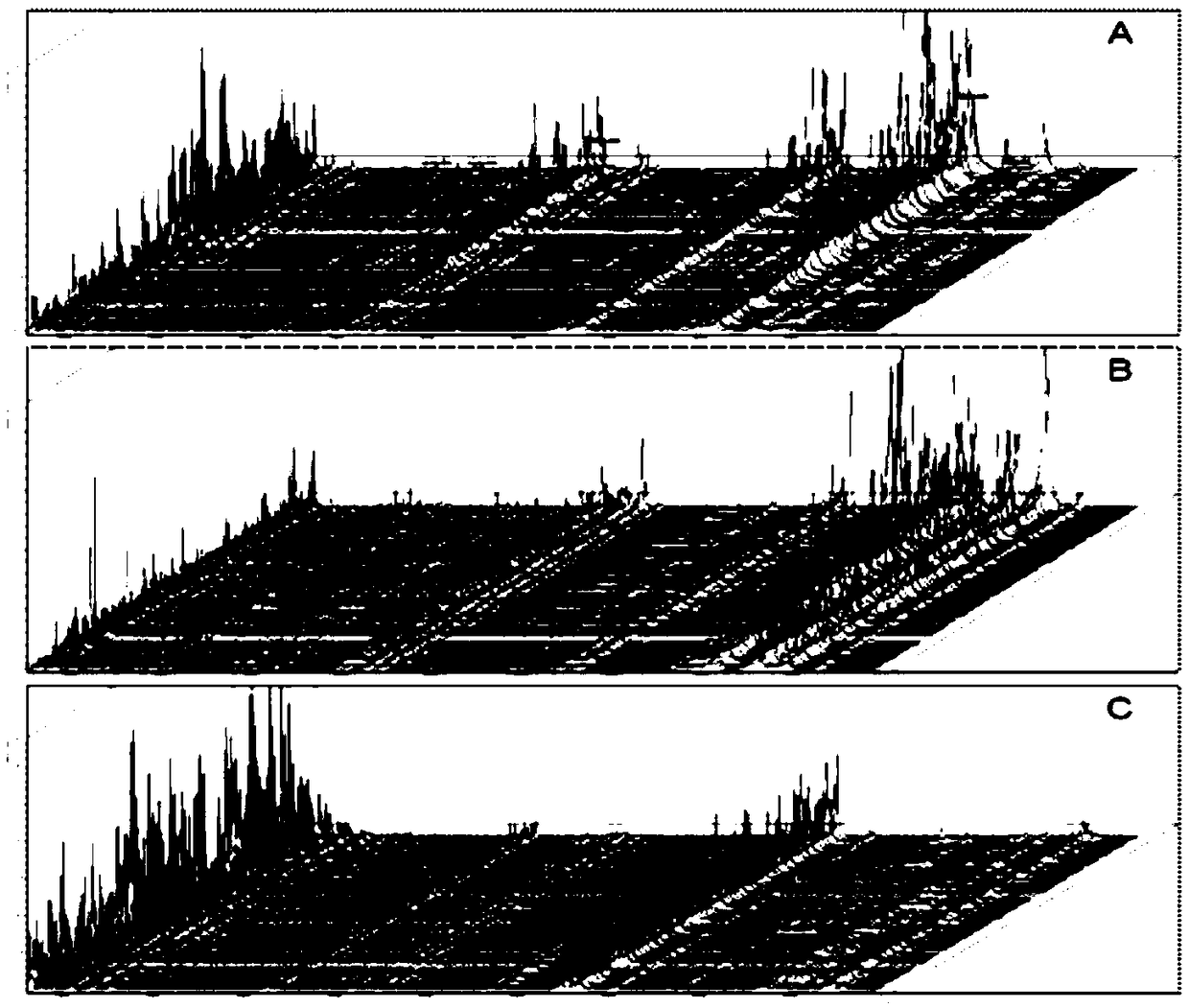
[0048] Collection of serum samples: Serum samples from cancer patients have been verified by the clinical gold standard. The sample information is shown in Table 1. Serum samples from 24 colorectal cancer patients, 24 liver cancer patients and 24 healthy people were collected; the male to female ratio of colorectal cancer patients in the samples was 15:9, the average age was 56, and the age range Between 20 and 72; the male to female ratio of liver cancer patients is 13:11, the average age is 60, and the age range is between 38 and 81; the male to female ratio of healthy people is 12:12, the average age is 58, and the age range is 51 Between -79.
[0049] Table 1 Serum sample information of patients with colorectal cancer, liver cancer and healthy people
[0050] sample source
Number of samples
male / female ratio
average age
age range
colorectal cancer patients
24
15 / 9
56
20-72
liver cancer patients
24
13 / 11
...
Embodiment 3
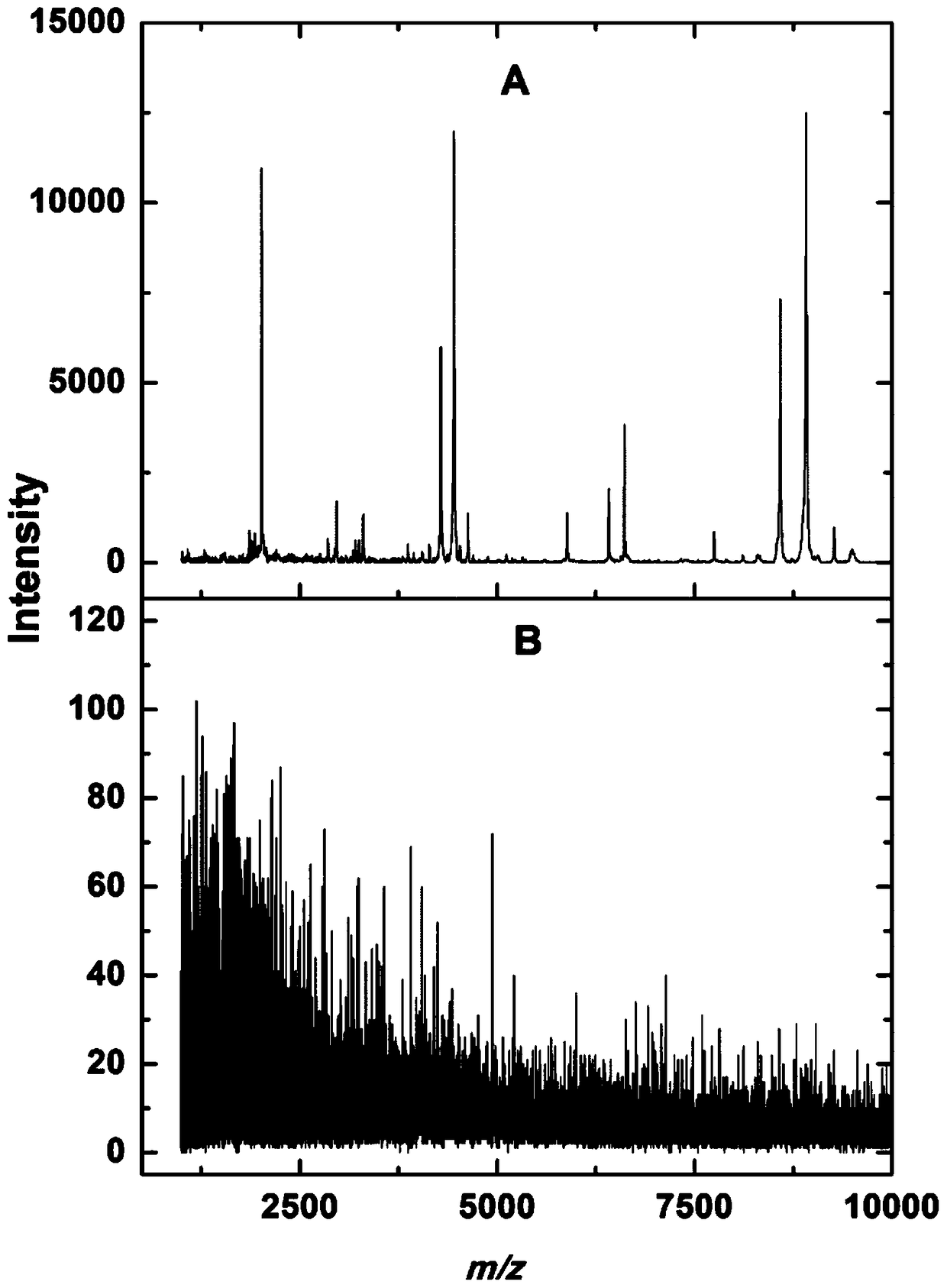
[0055] 1) Data processing: use Flex1n1lysis software to smooth the original spectrum and subtract baseline processing to eliminate background influence and noise interference, and then export the list of protein molecular weights. Perform peak alignment and normalization in M1TL1B, and use the t-test to select the three surface-modified porous silicon particles to detect the difference between healthy people and colorectal cancer patients, healthy people and liver cancer patients, and liver cancer patients and colorectal cancer patients. There were differences (p<10-7) between peaks; further screening finally obtained 8 peaks as characteristic values, m / z were 495.27, 1894.94, 3213.78, 4048.97, 6650.63, 8145.10, 8619.19, 9389.30.
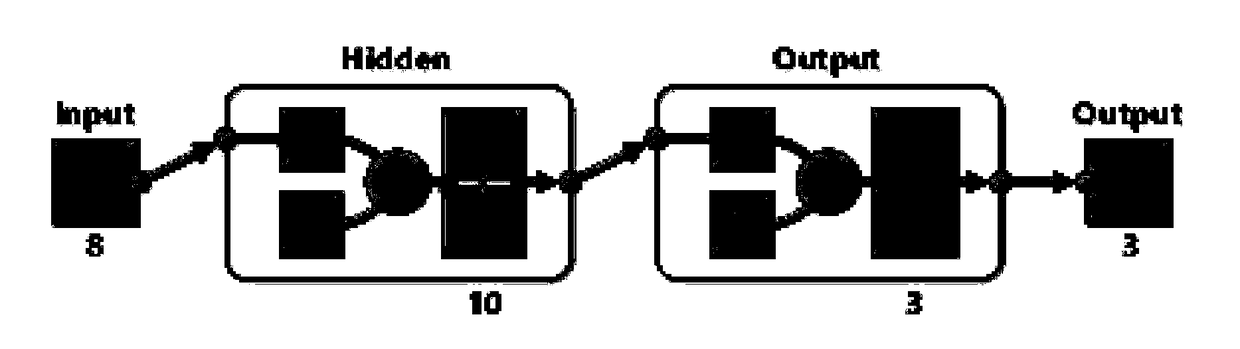
[0056] 2) Artificial neural network to establish a predictive model: Combine the generated characteristic peak data with artificial neural network software to generate a predictive model to diagnose colorectal cancer. The selected 8 peaks are used a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Aperture | aaaaa | aaaaa |
| Thickness | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com