Method for directionally knocking out miRNA (micro Ribonucleic Acid) of paddy rice
A rice and carrier technology, applied in the field of plant genetic engineering, can solve the problems of large blindness, difficulty in obtaining mutants, and difficult realization of miRNA, and achieve the effect of simple operation and high knockout rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
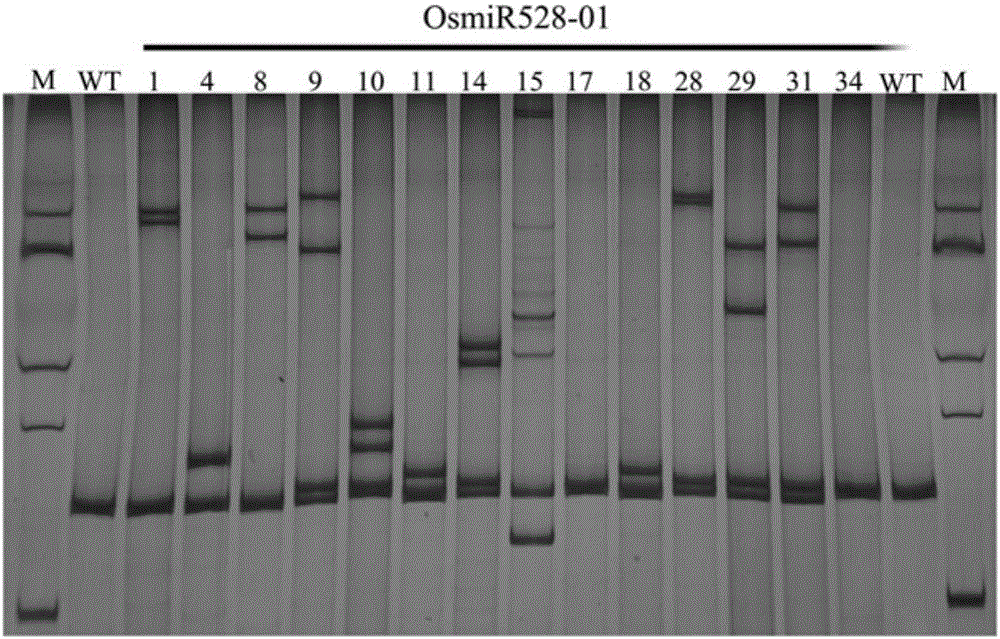
[0039] Example 1 Using the method of the present invention to create a single miRNA mutant (taking rice OsmiR528 as an example)
[0040] 1. Vector construction of CRISPR-Cas9-miRNA mutants
[0041] (1) Primer design
[0042] Primers were designed according to the recognition and cutting rules of the target site by CRISPR / Cas9. For the precursor genome sequence of rice OsamiR528 (GenBank number: GQ419957.2), the primers designed were OsmiR528-sgRNA1-F: 5'-gtgtGAAGGGGCATGCAGAGGAGC-3'; OsmiR528-sgRNA1-R: 5'-aaacGCTCCTCTGCA TGCCCCTTC-3'.
[0043] (2) Primer annealing
[0044] Dilute the upstream and downstream primers of each target site 10 times, take 10 μL each, denature at 98°C for 5 minutes, cool naturally, and dilute the annealed product 20 times for use.
[0045] (3) Digestion, gel recovery, connection
[0046] The backbone vector used in the experiment was pZHY988 ( figure 1 ), constructed by our laboratory, the construction process is as follows: first synthesize (syn...
Embodiment 2
[0090] Example 2 Using the method of the present invention to create multiple miRNA mutants (taking rice OsmiR397a and OsmiR97b as examples)
[0091] 1. Vector construction of CRISPR-Cas9-miRNAs mutants
[0092] (1) OsmiR397a and OsmiR397b double-site knockout vector construction method
[0093]The OsmiR397 family has two members, OsmiR397a (GenBank number: AP014962:28489785...28489898) and OsmiR97b (GenBank number: XM_015769221). In order to obtain the OsmiR397 knockout mutant, a two-site knockout mutation vector in which OsmiR397a and OsmiR97b were simultaneously knocked out was constructed. According to the precursor sequence of OsmiR397a and OsmiR97b, design and synthesize OsmiR397a-sgRNA1, gRNA scaffold, U6 terminator, U6 promoter, OsmiR397b-sgRNA1 sequence OsmiR397a-gRNA1-OsmiR397b-gRNA1-F and OsmiR397a-gRNA1-OsmiR397b-gRNA1- R (synthesized by Shanghai Yingjun Biotechnology Co., Ltd.). OsmiR397a-gRNA1-OsmiR397b-gRNA1-F:
[0094] gtgtGAGTGCAGCGTTGATGAACAAGTTTTAGAGCTAG...
Embodiment 3
[0105] Example 3 Using the method of the present invention to create miRNA large fragment deletion mutants (taking rice OsmiR408 as an example)
[0106] 1. Vector construction of CRISPR-Cas9-miRNAs mutants
[0107] (1) OsmiR408-sgRNA1 and OsmiR408-sgRNA2 double-site knockout vector construction method
[0108] When considering knocking out all or most of the precursor miRNAs or when it is difficult to find a suitable PAM site near some mature miRNA precursors, it is necessary to consider the strategy of using double-site knockout to create large fragment deletions.
[0109] According to the precursor sequence of OsmiR408, the sequences OsmiR408-gRNA1-gRNA2-F and OsmiR408-gRNA1-gRNA2-R containing sgRNA1, gRNA scaffold, U6 terminator, U6 promoter, and sgRNA2 were designed and synthesized (submitted by Shanghai Yingjun Biotechnology Co., Ltd. Ltd Synthetics).
[0110] OsmiR408-gRNA1-gRNA2-F:
[0111] gtgtGATGAGGCAGAGCATGGGATGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com