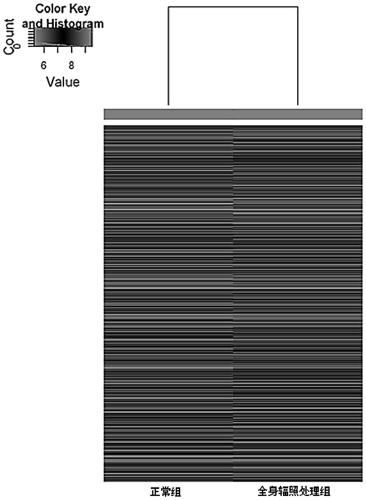
A leukemia-related circular circRNA-016901 gene and its use
A gene, circular technology, applied in the field of circular circRNA-016901 gene, can solve the problems of transplantation failure, unclear hematopoiesis and its mechanism of action, affecting HSCs engraftment and transplantation efficacy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Amplification of circular circRNA-016901 gene primer pair
[0022] According to the cDNA sequence shown in SEQ ID NO:1 of the circular circRNA-016901 gene, primers for amplification were designed, and the specific sequences of each primer are shown in Table 1.
[0023] Table 1 Circular circRNA-016901 Gene Amplification Primer Sequence
[0024] Primer name Sequence (5'→3') SEQ.ID F ACAGCGCTACACTTGTTCCGA 2 R GACGATGCTATCCAGGAGAGGT 3
[0025] After the above primers were artificially synthesized, they were used as a primer pair for amplifying the cDNA sequence of the circular circRNA-016901 gene.
Embodiment 2
[0026] Example 2: Preparation of total RNA in the sample
[0027] According to the RNA extraction steps of TRIZOL method, the total RNA in the bone marrow stromal cell samples of the control group and the whole body irradiation treatment group was extracted. A brief description is as follows:
[0028] 1. Homogenate
[0029] According to the instructions of the TRIZOL method, the sample was homogenized after adding TRIZOL reagent according to the number of cells;
[0030] 2. Two-phase separation
[0031] After incubating the homogenized sample at 15-30°C for 5 minutes, add 0.2ml of chloroform to every 1ml of TRIZOL reagent homogenized sample, and tightly cap the tube. Shake the tube vigorously by hand for 15 seconds, and incubate at 15 to 30°C for 2 to 3 minutes. Centrifuge at 12,000 x g for 15 min at 4°C. After centrifugation, the mixed liquid will be divided into the red phenol chloroform phase of the lower layer, the colorless aqueous phase of the middle layer and the u...
Embodiment 3
[0041] Embodiment 3: the synthesis of total cDNA sequence
[0042] The total RNA that passed the quality test was synthesized according to the method described below. The reagents used in the synthesis and their manufacturers are as follows:
[0043] RNase inhibitor (Epicentre); SuperScriptTM III Reverse Transcriptase (Invitrogen); 5×RT buffer (Invitrogen); 2.5mM dNTP mixture (dATP, dGTP, dCTP and dTTP each 2.5mM) (HyTest Ltd); Jun Biotechnology Co., Ltd.)
[0044] The thermocycler used for cDNA synthesis was Gene Amp PCR System 9700 (Applied Biosystems).
[0045] The specific operation method is as follows:
[0046] 1. Prepare the annealing mixture
[0047] RNA 800ng
[0048] 0.5ug / ul Random (N9) 1μl
[0049] dNTPs Mix (2.5mM) 1.6μl
[0050] Add RNase-free H 2 O to a total volume of 14.5 μl; the mixture was placed in a water bath at 65°C for 5 minutes, and placed on ice for 2 minutes.
[0051] 2. After short centrifugation, add RT reaction solution to the centrifuge tu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com