Meloidogyne incognita Chitwood prevention and control related gene Misp12 and use thereof
A technology of root-knot nematode incognita and sequence, which is applied in application, nematicides, genetic engineering, etc., can solve the problems of unknown function of Misp12 and no homologous genes retrieved, so as to achieve environmental protection and safety off-target effects, reduce the number of invasions, and avoid The effect of off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
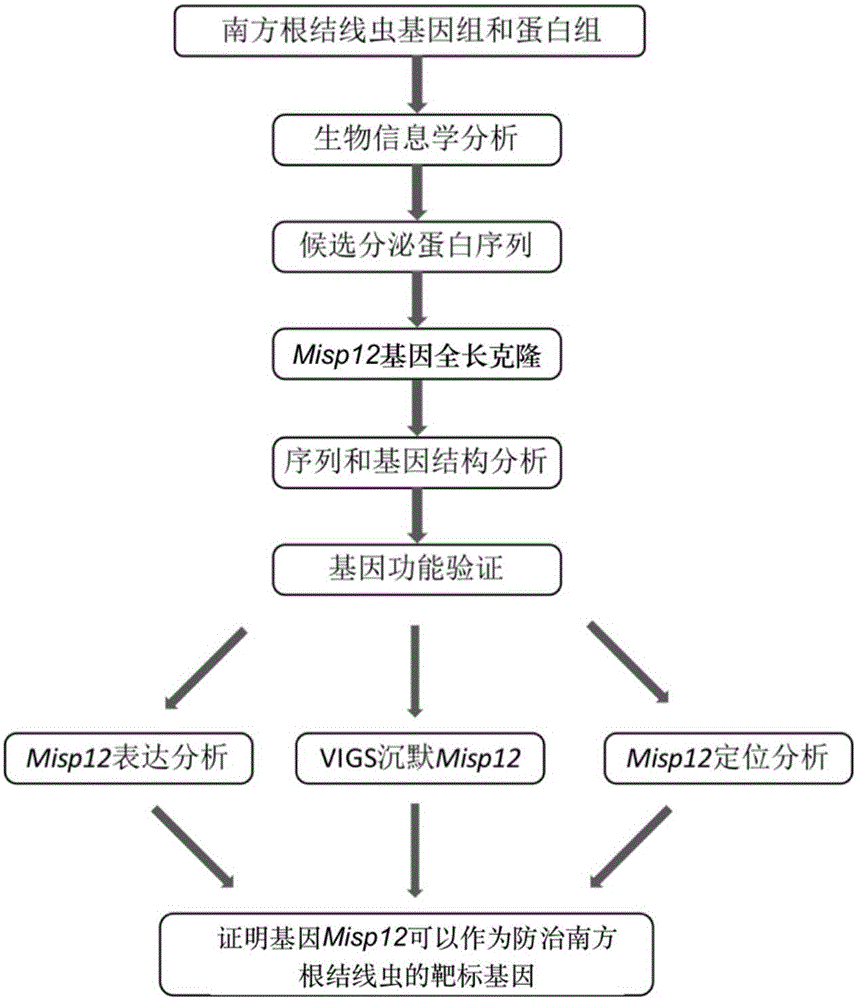
[0033] Cloning of Misp12 gene related to developmental pathogenicity of Meloidogyne incognita
[0034] The applicant obtained the candidate secreted protein sequence by analyzing and comparing the genome data and proteome data of Meloidogyne incognita through bioinformatics technology, and isolated and cloned the gene Misp12 through molecular biology technology. Through genome database and proteome database sequence comparison, it is found that Misp12 is highly specific and conserved in M. The growth and development of root-knot nematodes may be related to the pathogenic process and play an important role ( figure 1 ).
[0035] 1.1 Bioinformatics analysis of genome and proteome data of M. incognita
[0036] Download the genome and proteome data of M. incognita through NCBI ( http: / / meloidogyne.toulouse.inra.fr ). Use SignalP 3.0 and THMM2.0 to analyze the signal peptide structure and transmembrane structure of its proteome to obtain secretome proteins, and then use variou...
Embodiment 2
[0044] Bioinformatic analysis of Misp12 protein and its homologous proteins in other fungi
[0045] The amino acid sequence putatively encoded by the Misp12 gene was analyzed through the NCBI BLAST database and the Swissprot database, and it was found that this sequence only exists in Meloidogyne incognita, and there are no corresponding homologous sequences and homologous proteins in other animals, plants, fungi and bacteria. Motif analysis of the protein using MotifScan shows that it contains 2 predicted N-glycosylation sites, 2 predicted protein kinase C phosphorylation sites, 1 cAMP and cGMP dependent phosphorylation site and 1 N- Myristoylation site ( Figure 4 ).
Embodiment 3
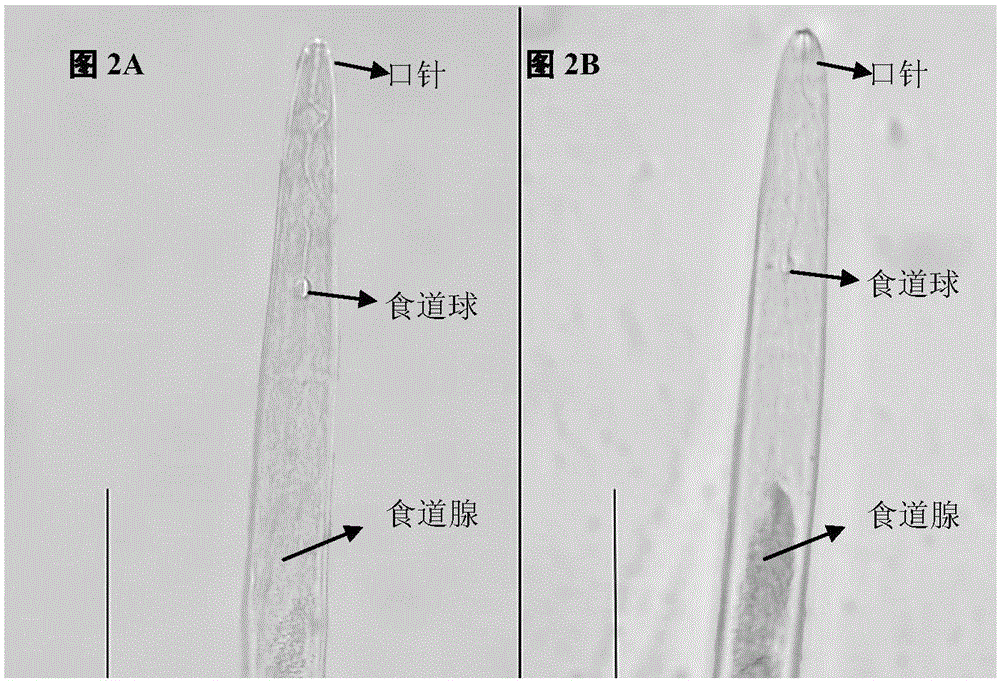
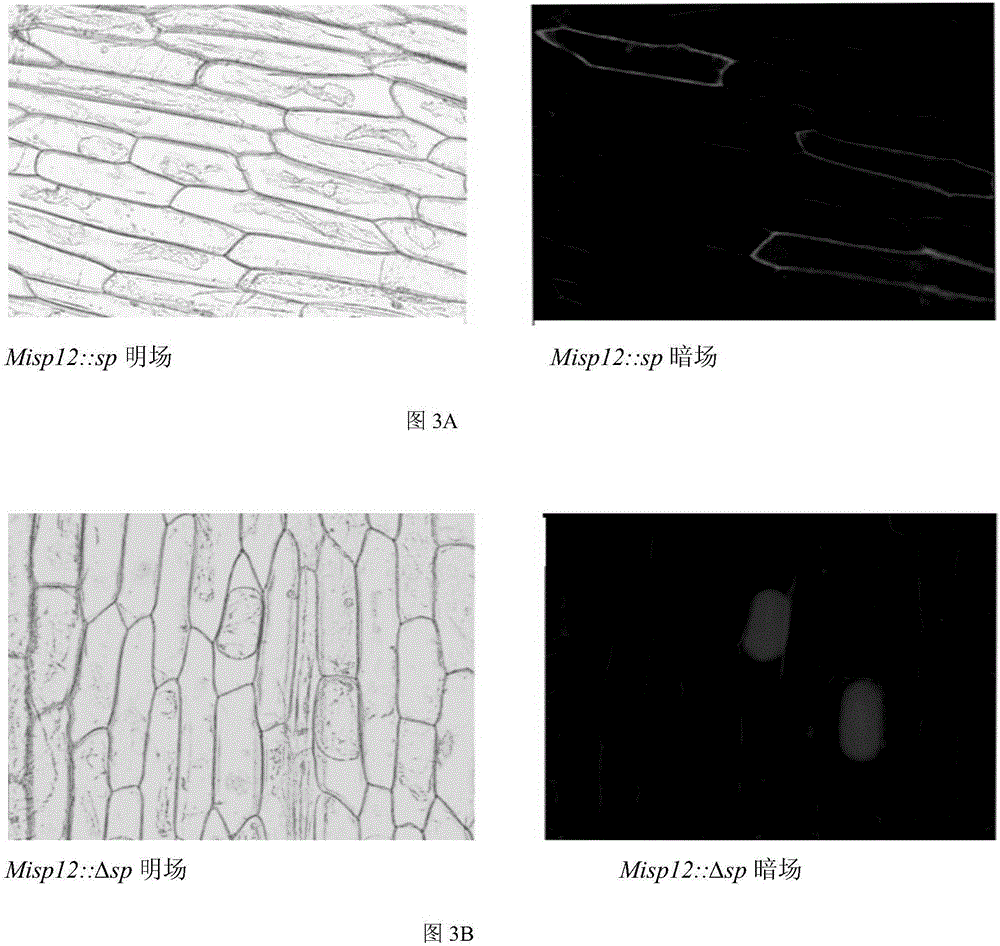
[0047] Localization of Misp12 gene in Meloidogyne incognita and its protein localization in plant cells
[0048] The expression position of Misp12 gene in Meloidogyne incognita was detected by in situ hybridization, and Misp12 gene fused with gfp gene was transferred into onion epidermal cells to observe its localization in plant cells.
[0049] 3.1 In situ hybridization test
[0050] 3.1.1 Solution preparation
[0051] (1) M9Buffer: 22mM KH 2 PO 4 , 42.3mM NaHPO 4 , 85.6mM NaCl, 1mM MgSO 4 . 7H2O
[0052] (2) Fixative: 3% paraformaldehyde, prepared with 1×M9Buffer
[0053] (3) Proteinase K: 2mg / ml, prepared with 10mM Tris-HCl (PH 7.5)
[0054] (4) For the preparation of other hybridization reagents, refer to the instructions of the Digoxigenin Labeling Kit (purchased from Roche)
[0055] 3.1.2 Digoxigenin-labeled DNA single-strand probe synthesis
[0056] The single-stranded sense probe and antisense probe of the target gene were synthesized by using the digoxin lab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com