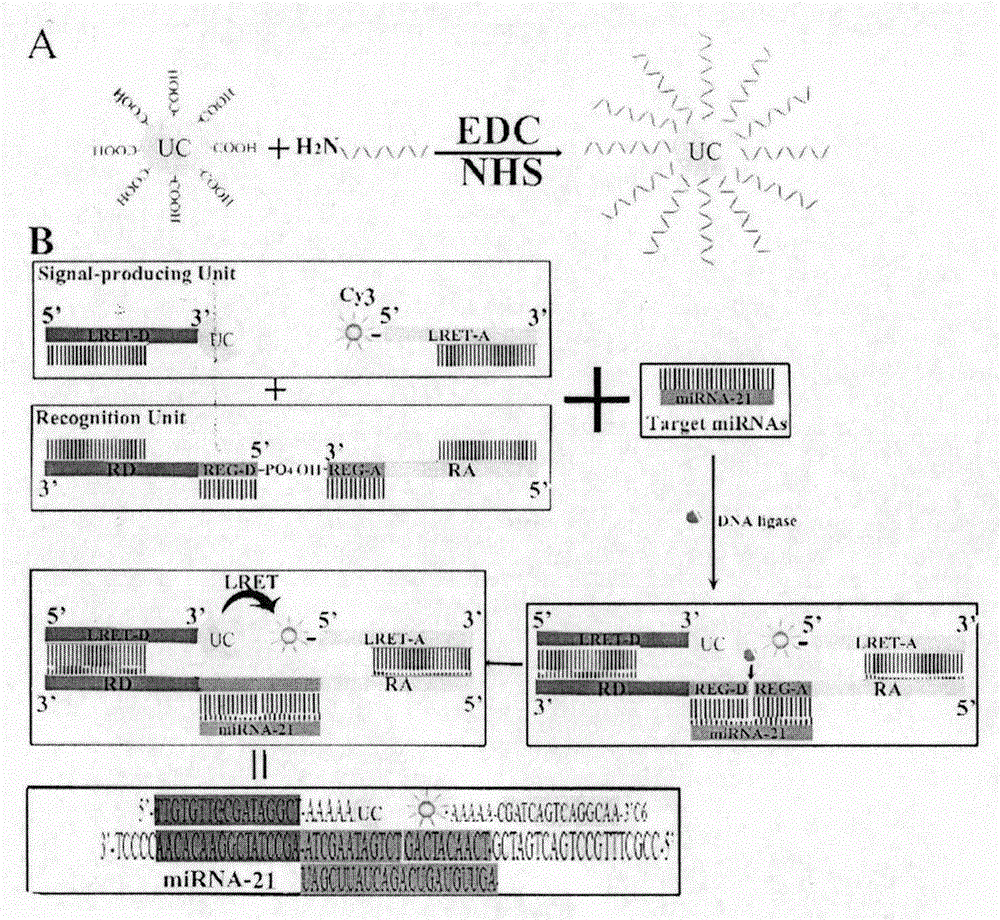
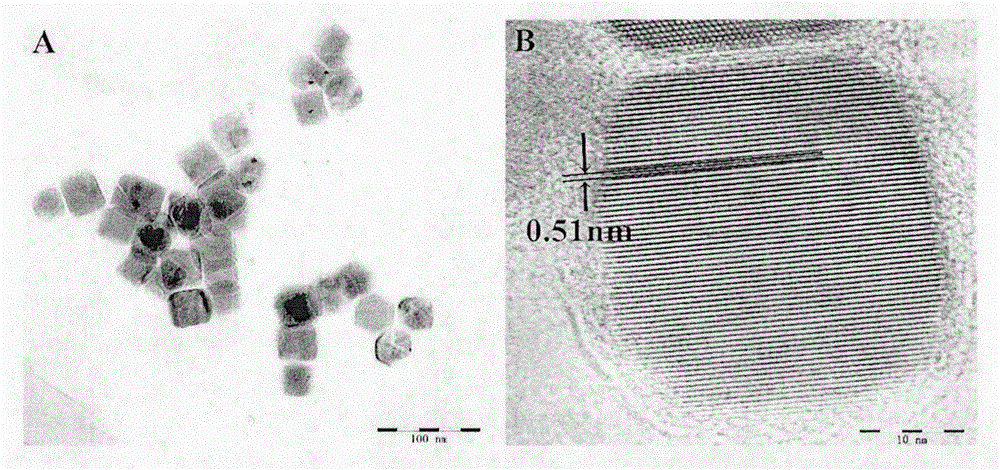
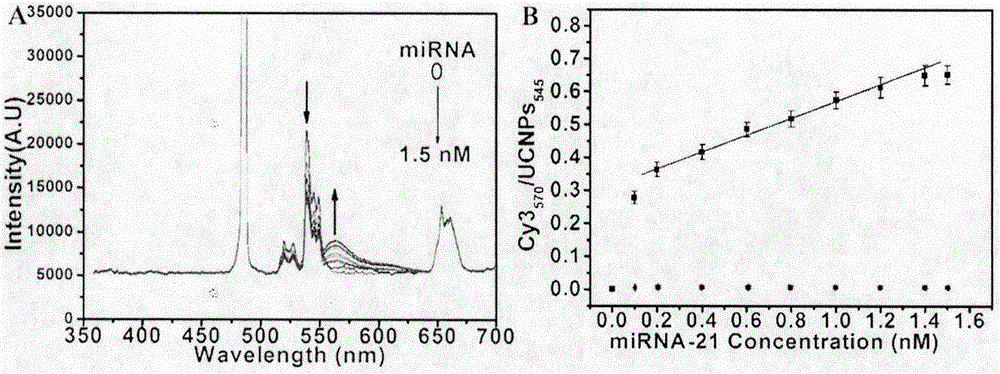
Double-recognition quantitative detection method for microRNA
A quantitative detection method and quantitative detection technology, applied in the fields of biotechnology and clinical medical diagnosis, can solve the problems of labor consumption, time-consuming detection of miRNA, technical difficulty in large-scale expression analysis, etc., to avoid background fluorescence and poor detection performance Falling, good stability effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Upconverting Nanoparticles (NaYF 4 : Er 3+ , Yb 3+ ) labeled oligonucleotide (LRET-B)
[0024]Experimental materials: bovine serum albumin, batch number 140318, Shanghai Aladdin Reagent Co., Ltd.; 1-ethyl-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), batch number 090M14531V, Shanghai Aladdin Reagent Co., Ltd. Latin Reagent Co., Ltd.; Succinimide (NHS), batch number MKBG7914V, Shanghai Aladdin Reagent Co., Ltd.; Ethanol, batch number: 14021710247, purchased from Nanjing Chemical Reagent Co., Ltd. The oligonucleotide sequence (eg 5'-TTGTGTTCCGTAGGCT-AAAAAAAA 3'C7-NH2) from the 5' end is the specific sequence that is complementary to the recognition unit, the sequence that controls the length of the oligonucleotide, and the 3' end modified C7 amino The sequence composition, in which TTGTGTTCCGATAGGCT is a specific sequence that is complementary to the recognition unit, varies with the detected miRNA. The oligonucleotide sequence is:
[0025] UC-L...
Embodiment 2
[0041] Example 2: Quantitative detection of miRNA-21 based on dual recognition microRNAs
[0042] Experimental material: ethanol, batch number: 14021710247, purchased from Nanjing Chemical Reagent Co., Ltd. HEPES buffer, batch number: E607018, purchased from Shanghai Sangon Biological Reagent Co., Ltd.; 5′ adenosine triphosphate disodium salt trihydrate (ATP), batch number: A600020, purchased from Shanghai Sangon Biological Reagent Co., Ltd.; single stranded salmon sperm sperm DNA, batch number: C640031, was purchased from Shanghai Sangon Biological Reagent Co., Ltd.; RNA ligase, batch number: B101801, was purchased from Shanghai Sangon Biological Reagent Co., Ltd.
[0043] The oligonucleotide sequence is:
[0044] UC-LRET-B: 5’-TTGTGTTCCGATAGGCT-AAAAAAAA 3’ C7-NH 2 ;
[0045] UC-LRET-A: Cy3-5’ AAAAAAAA-CGATCAGTCAGGCAA-3’ C6;
[0046] the adaptor oligos (for miRNA-21):
[0047] (RD-REG-A): 3’ TCCCCAACACAAGGCTATCCGA-ATCGAATAGTCT-5’-PO 4
[0048] (RD-REG-D): 3’ OH-GACTACA...
Embodiment 3
[0052] Example 3: Quantitative detection of miRNA-125a based on dual recognition microRNAs
[0053] Experimental material: ethanol, batch number: 14021710247, purchased from Nanjing Chemical Reagent Co., Ltd. HEPES buffer, batch number: E607018, purchased from Shanghai Sangon Biological Reagent Co., Ltd.; 5′-adenosine triphosphate disodium salt trihydrate (ATP), batch number: A600020, purchased from Shanghai Sangon Biological Reagent Co., Ltd.; single stranded salmon sperm DNA, batch number: C640031, purchased from Shanghai Sangon Biological Reagent Co., Ltd.; RNA ligase, batch number: B101801, purchased from Shanghai Sangon Biological Reagent Co., Ltd.;
[0054] The oligonucleotide sequence is:
[0055] UC-LRET-B: 5’-TTGTGTTCCGATAGGCT-AAAAAAAA 3’ C7-NH 2 ;
[0056] UC-LRET-A: Cy3-5’ AAAAAAAA-CGATCAGTCAGGCAA-3’ C6;
[0057] the adaptor oligos (for miRNA-125a):
[0058] (RD-REG-A): 3’ TCCCCAACACAAGGCTATCCGA-AGGGACTCTGGG-5’-PO 4
[0059] (RD-REG-D): 3' OH-AAATTGGACACT-GCT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com