Sequencing library and preparation and application thereof
A sequencing library and sequencing technology, applied in the field of sequencing library and its preparation and application, can solve the problem that the accuracy of DNA sequencing cannot meet the actual needs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
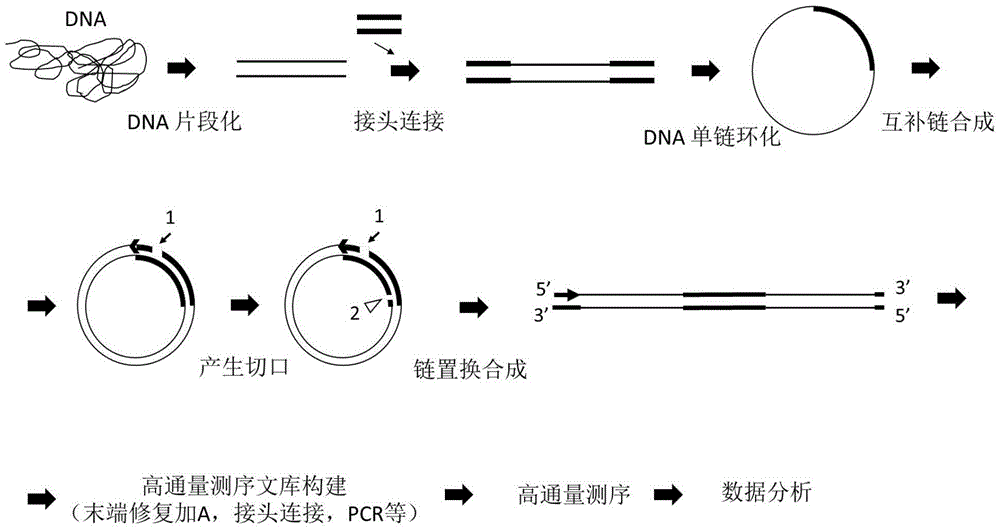
[0094] Example 1: Construct a whole-genome DNA library (Illumina platform) according to the above scheme 1 (double gap scheme)
[0095] 1) DNA fragmentation
[0096] Instruments and reagents used:
[0097] Ultrasonic interrupter: Covaris: S2 Focused-ultrasonicator
[0098] Interruption tube: Covaris Microtube 6x16mm, catalog#: 520045
[0099] QIAGEN MinElute Gel Extraction Kit(250), Catalog#:28606
[0100] Takara 20bp DNA Ladder (Dye Plus), Takara Code, 3420A
[0101] 5 μg of purified PhiX174 genomic DNA was broken into 150-200 bp with an ultrasonic breaker (Covaris S2Focused-ultrasonicator) (Intensity: 5, Duty Cycle: 10%, Cycles per Burst: 200, Temperature: 4°C, time: 60s ,number ofcycles:5), the interruption system is 50μl.
[0102] 4% agarose gel electrophoresis (80V, 70min; 1×TAE), gel cutting and recovery (QIAGEN MinEluteGel Extraction Kit) 60-90bp fragment (Takara 20bp DNA Ladder), the recovery steps are detailed in the QIAGEN MinElute Gel Extraction Kit manual.
...
Embodiment 2
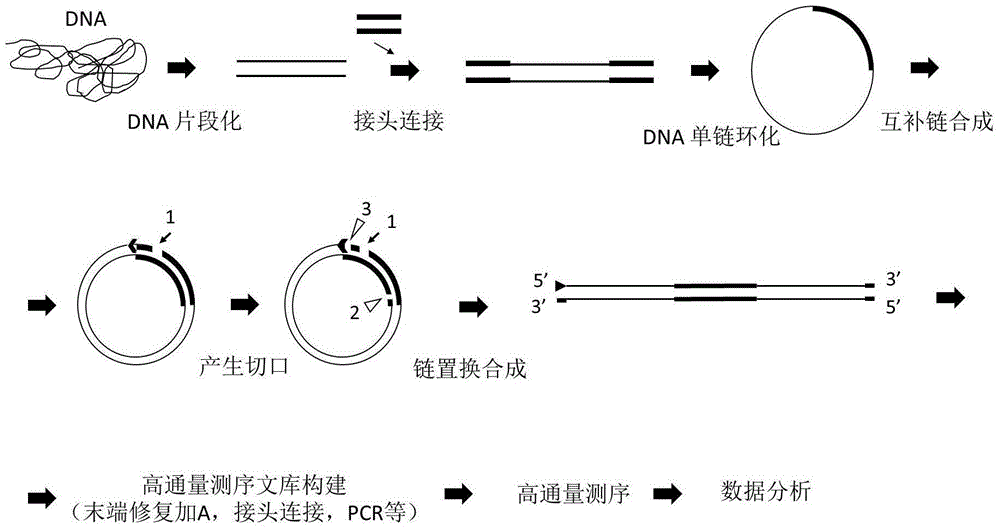
[0185] Example 2: Construction of a genome-wide DNA library according to the above-mentioned scheme two (here, the three-gap scheme is taken as an example)
[0186] (1) DNA fragmentation, adding A to the end, connecting adapters, single-strand circularization steps are the same as in Example 1.
[0187] (2) Complementary strand synthesis
[0188] New England Biolabs: Klenow Fragment (3'→5'exo-), Catalog#:M0212S
[0189] New England Biolabs:USER TM Enzyme, Catalog #: M5505S
[0190] NEB buffer 4: 2μl
[0191] primer (UO-p1-2, 10uM): 1μl
[0192] DNA: 15.8 μl
[0193] 95°C for 3 minutes, and immediately placed on ice for 3 minutes.
[0194] When done add:
[0195] 2.5mM dNTPs: 0.5μl
[0196] 100X BSA: 0.2 μl
[0197] Klenow Fragment (3'→5'exo-): 1μl
[0198] Total 20μl
[0199] 20°C for 30 minutes, 75°C for 20 minutes.
[0200] USER TM Enzymes: 1μl
[0201] 37°C, 30min, 50°C, 5min, immediately put on ice.
[0202] The product was purified using Agencourt AMPure ...
Embodiment 3
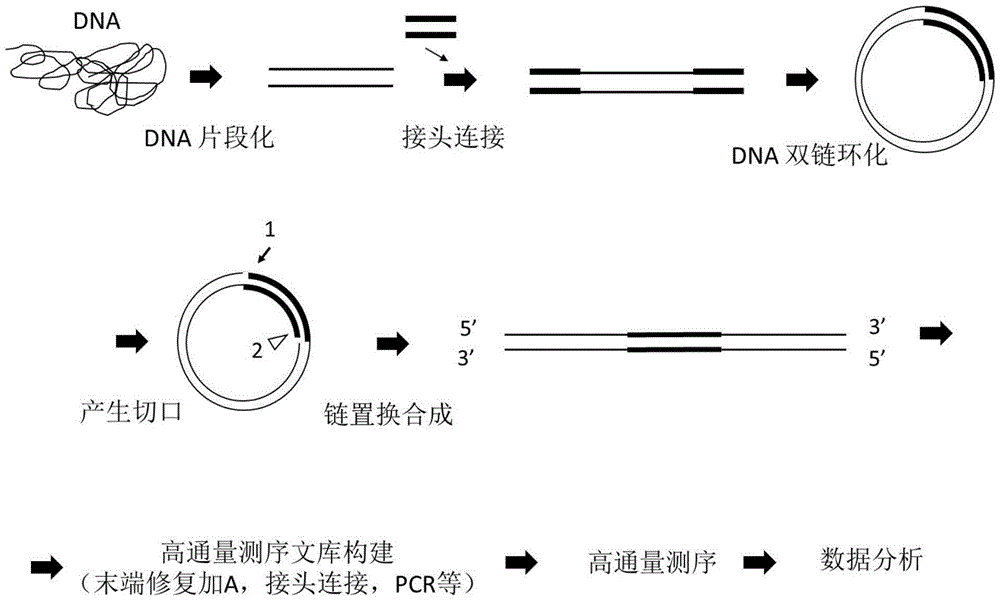
[0205] Example 3: Construction of a whole genome DNA library according to the above-mentioned scheme three (double-stranded circularization scheme, the linker contains a notch to be cut)
[0206] (1) DNA fragmentation (~700bp, fragmentation conditions: duty cycle: 5%, intensity: 3, cyclesper burst: 200, time: 75s)), fill in the end and add A, connect the adapter with the same example 1, the adapter sequence It is: UO-A2, which is formed by annealing the following two sequences:
[0207] 5'-AGCACGTACGACTGAUCT-3' (SEQ ID NO: 5)
[0208] 5'-pGATCAGTCGTACGTGCT-3' (SEQ ID NO: 6)
[0209] (2) terminal phosphorylation
[0210] 44μl DNA, 10U T4PNK (T4Polynucleotide Kinase, NEB, M0201S), 50mMTris-HCl pH 7.5, 10mM MgCl2, 1mM ATP, 10mM DTT, 37°C for 30min, the product was purified with 1XAmpure XP magnetic beads.
[0211] (3) Double-strand cyclization
[0212] Quick Ligation Module(NEB,E6056S)
[0213] DNA: 35 μl
[0214] T4quick ligase: 5 μl
[0215] 5Xligase buffer: 10μl
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com