Primers, probes, kit and method for detecting proteus mirabilis
A technology of Proteus mirabilis and a kit, which is applied in the field of microbial detection, can solve the problems of easy generation of false positives, long cycle and low efficiency, and achieves the effects of good specificity, low cost and convenient use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1 Proteus mirabilis detection kit
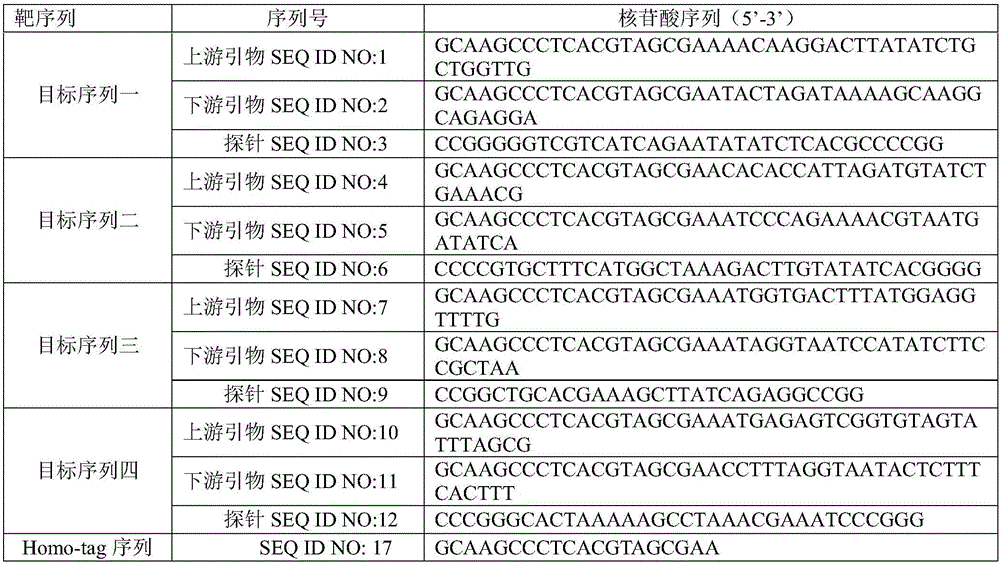
[0062] 1. Design primers and probes for target sequence one obtained by whole gene sequencing, target sequence two, target sequence three and target sequence four, synthesize the nucleotide sequences and Homo-tag sequences of its primers and probes (as shown in Table 1 DABCYL is connected to the 3' end of each probe sequence, and the fluorophore corresponding to the 5' end of each probe sequence is: SEQ ID NO: 3 corresponds to HEX, SEQ ID NO: 6 corresponds to ROX, and SEQ ID NO: 6 corresponds to ROX. ID NO: 9 corresponds to CY5 and SEQ ID NO: 12 corresponds to FAM.
[0063] Each of the above primers and probes was prepared as a storage solution with a concentration of 0.2 μM, and the Homo-tag sequence was prepared as a storage solution with a concentration of 0.6 μM.
[0064] 2. Separately prepare PCR buffer, MgCl 2 solution, dNTP solution, rTaq enzyme solution; among them, MgCl 2 The concentration is 3mM, the dNTP conce...
Embodiment 2
[0066] The detection and analysis of embodiment 2 Proteus mirabilis
[0067] 1. Sample nucleic acid extraction: Take about 0.2g or 200μl of stool samples from patients with diarrhea caused by clinical Proteus mirabilis and healthy people respectively, add them to 1.5ml EP tubes, add the sample processing solution (PBS buffer or normal saline), Shock 3 times, 10 seconds each time. Let it stand for 10 minutes, then centrifuge at 8000rpm for 5 minutes, draw the supernatant and directly boil it for nucleic acid extraction to obtain 10-100ng / μl nucleic acid template.
[0068] 2. Fluorescent quantitative PCR reaction:
[0069] Take the detection kit prepared in Example 1, add 5.0 μl of the nucleic acid template obtained in the above step 1 to each PCR reaction tube, and the components of the 25 μl reaction system in each PCR reaction tube are as follows:
[0070]
[0071]
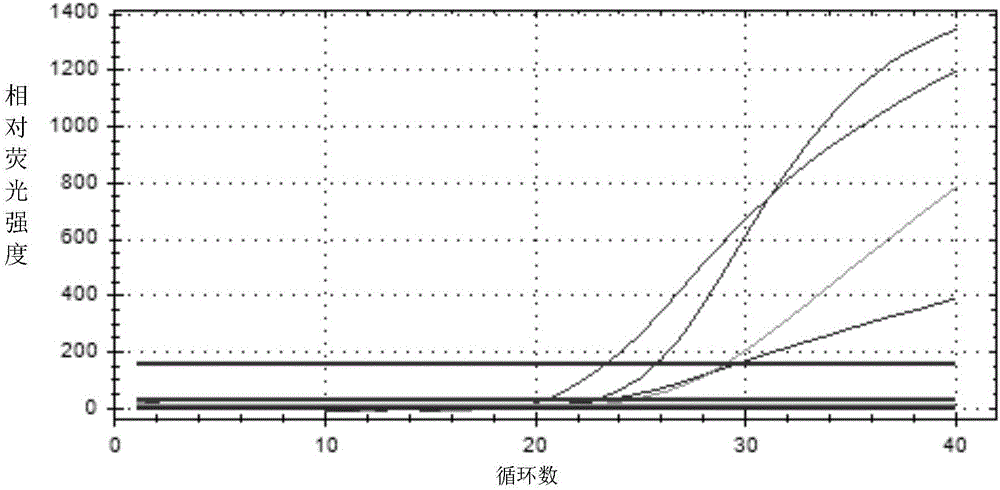
[0072] Then, send the PCR reaction eight tubes into a fluorescent PCR instrument (staragene-3005) for ...
Embodiment 3
[0079] Embodiment 3 specificity analysis
[0080] The applicant simultaneously obtained 194 strains involving bacteria such as Enterobacter, cocci, and Vibrio, and used the detection kit of Example 1 to carry out specific analysis experiments, and used the positive strain of Proteus mirabilis as a control. The situation of the detected strains is shown in Table 3. The test results were all negative. The results of this example show that: the primers and probes in the kit provided in Example 1 of the present invention have good specificity, and no non-specific amplification occurs, thereby avoiding false positives caused by non-specificity.
[0081] table 3
[0082]
[0083]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com