Nucleic acid and application thereof
A nucleic acid and target gene technology, which is applied in the direction of nucleic acid carrier, the use of carrier to introduce foreign genetic material, and cells modified by introducing foreign genetic material, etc., to reduce the difficulty of synthesis and increase the effect of down-regulating miRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Design of miRNA functional down-regulation sequence.
[0036] After clarifying the miRNAs that need to be functionally down-regulated, the binding sequences of the target genes that have been reported or can be predicted are searched, and the binding sequences of the target genes are connected in series.
[0037] Taking miR-1915-3p, which has a high CG content and is difficult to down-regulate functionally, as an example, the individual binding site sequences of target genes are shown in Table 1.
[0038] Table 1:
[0039]
[0040] The sequence designed to functionally down-regulate miR-1915-3p is as follows:
[0041]
[0042] The underline is the miRNA target gene binding site sequence, the underlined bold part is the miRNA target gene complementary pairing sequence, the bold italic part is the recognition site of EcoRI and EcoRV, respectively.
Embodiment 2
[0043] Example 2 Construction of miRNA functional down-regulation plasmid
[0044] 1. According to the design of Example 1, the sequence was synthesized.
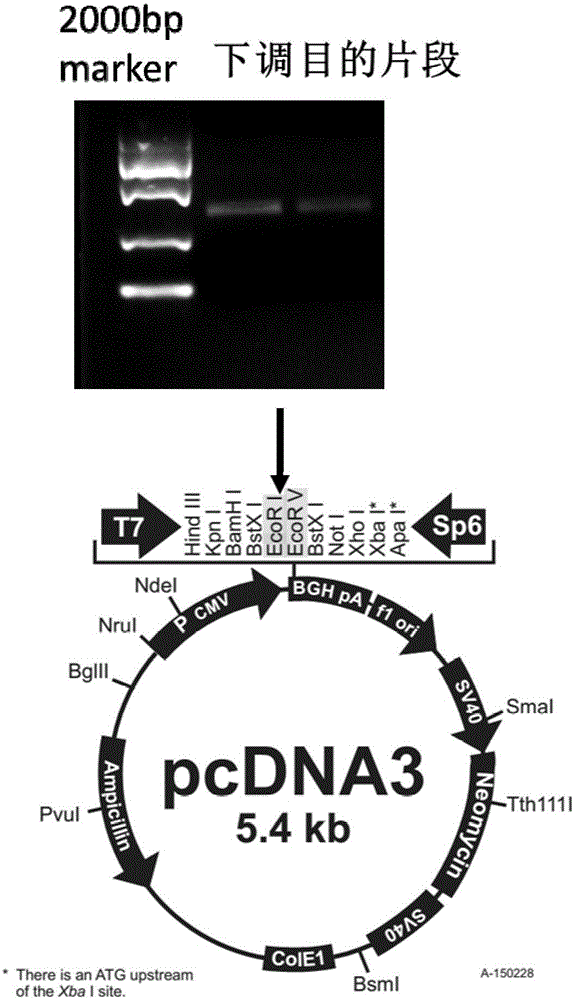
[0045] 2. The fragment synthesized in Example 1 was digested with EcoRI and EcoRⅤ by double enzyme digestion ( figure 1 The agarose gel electrophoresis diagram of the fragment synthesized in Example 1 after digestion with EcoRI and EcoR V), and ligate it into the target plasmid (the plasmid map is as figure 1 Shown).
[0046] Ligated into plasmid pCDNA3.1, restriction digestion system is shown in Table 2.
[0047] Table 2:
[0048] Composition Quantity Plasmid 4μg EcoRⅠ10IU EcoRⅤ10IU 10×Buffer 4μl water Add to 40μl
[0049] Mix the ingredients of the digestion system thoroughly and incubate at 37°C for 5 hours
[0050] 3. Purification and recovery
[0051] The digested product was subjected to agarose gel electrophoresis, and purified and recovered with a common DNA product purification kit.
[0052] 4. Connection of plasmids
...
Embodiment 3
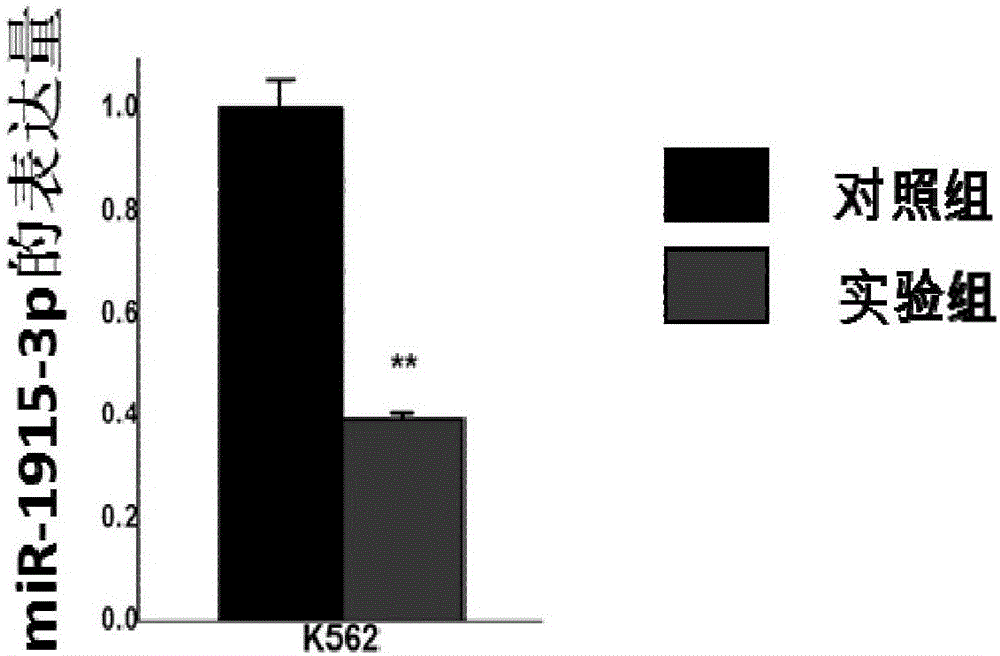
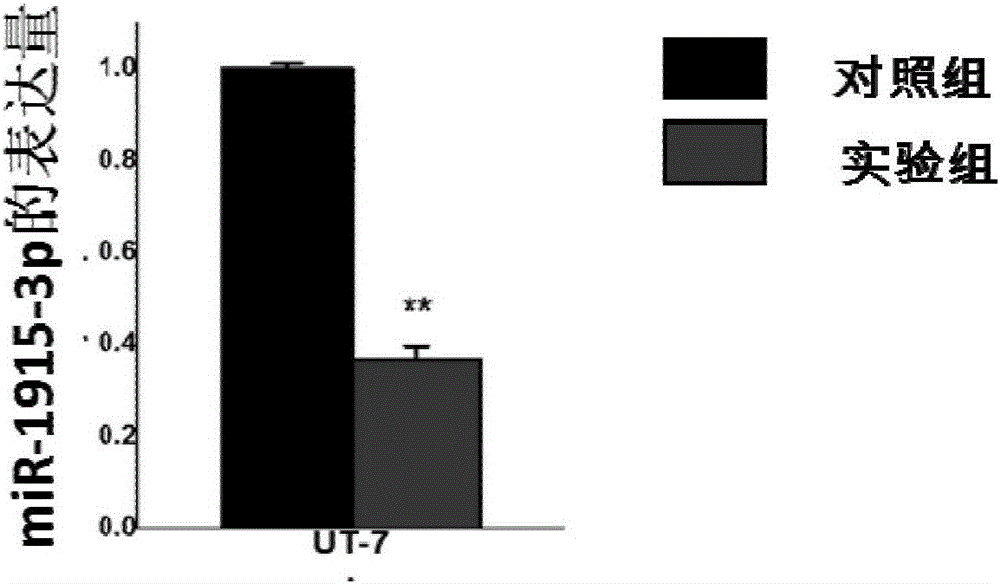
[0056] Example 3 Functional verification of miRNA functional down-regulation plasmid
[0057] 1. Establishment of a cell line that stably down-regulates miR-1915-3p
[0058] (1) Human leukemia cells (such as K562 cells or UT-7 cells) are cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), and passaged 1:5 every 3 days. Centrifuge the cells of the leukemia cell line, count them, and resuspend them in medium to a cell density of 3×10 5 Pcs / ml, add 1.5ml per well to the labeled 6-well plate;
[0059] (2) Take 4μg of purified plasmid and fill up to 250μl with Opti-MEM medium (EP tube 1), and mix; take another 1.5ml EP tube, add 240μl Opti-MEM medium and 10μl Lipofectamine 2000 (EP tube 2) ), flick and mix well, let stand at room temperature for 5 min;
[0060] (3) Mix the liquids in EP tubes 1 and 2 and mix gently. After standing at room temperature for 20 minutes, add the corresponding liquid to the cell suspension in the prepared 6-well plate drop by drop, shake...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com