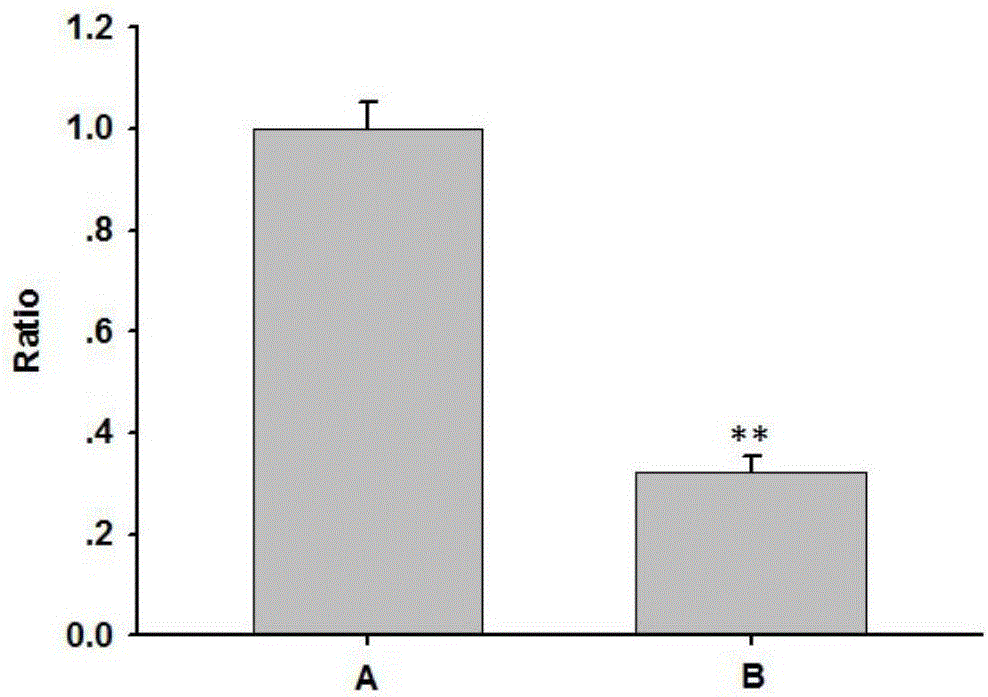
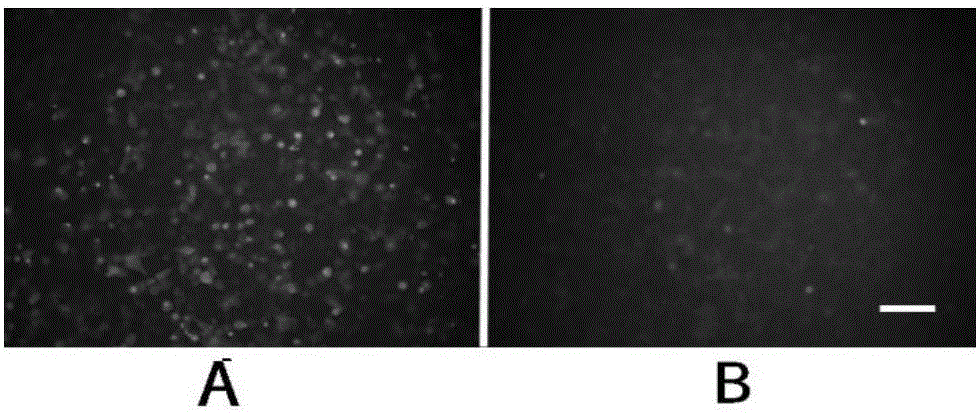
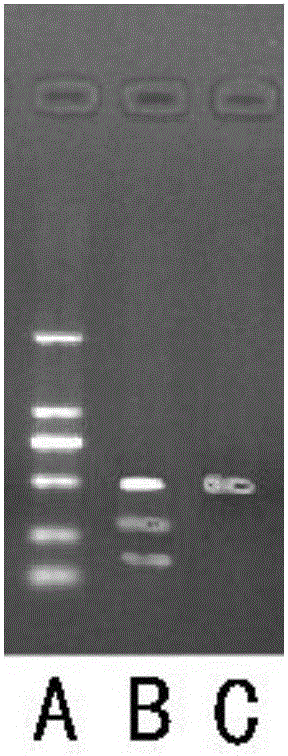
Gene knockout test kit and method for rapidly screening sgRNA
A gene knockout and kit technology, which is applied in the direction of retroRNA virus, genetic engineering, plant gene improvement, etc., can solve the problem of low activity of sgRNA-mediated genome modification, and achieve the effect of shortening the experimental cycle and cost, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] 1. Construction of pCAG-target-mCherry-Luc plasmid vector
[0065] The construction of the target gene pCAG-target-mCherry-Luc fluorescence-luciferase double reporter gene plasmid of sgRNA screening system in the present invention, wherein reagent A contains pCAG-mCherry-Luc fluorescence-luciferase double reporter gene plasmid, adopts 人工合成人源RPA3靶基因DNA CGCGGATCCGGTGCCGAGAAATCTGCCCCATAGACACCCGCGGGGCGCACAGTTTCAGTCGTCCGTGGGTTTCCCGCCAGCCGCAGTCTTGGACCATAATCATGGTGGACATGATGGACTTGCCCAGGTCGCGCATCAACGCCGGCATGCTAGCTCAATTCATCGACAAGCCTGTCTGCTTCGTAGGGAGGCTGGAAAAGATTCATCCCACCGGAAAAATGTTTATTCTTTCAGATGGAGAAGGAAAAAATGGAACCATCGAGTTGATGGAACCCCTTGATGAAGAAATCTCTGGAATTGTGGAAGTGGTTGGAAGAGTAACCGCCAAGGCCACCATCTTGTGTACATCTTATGTCCAGTTTAAAGAAGATAGCCATCCTTTTGATCTTGGACTTTACAATGAAGCTGTGAAAATTATCCATGACTTCCGAATTCCGG(SEQ ID NO:2),靶基因DNA的两端含有BamHI及EcoRI酶切位点,其中5端为BamHI,3端为EcoRI;pCAG-mCherry-Luc质粒和 The artificially synthesized target gene DNA was double-digested with BamHI and EcoRI, mixed, ligated with T4 DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com