Real-time fluorescence PCR method for quantitatively detecting Pseudomonas syringae pv. Actinidiae
A technology of canker bacteria and real-time fluorescence is applied in the field of plant pathogen detection to achieve high sensitivity, improve detection efficiency and sensitivity, and prevent the expansion of the affected area
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
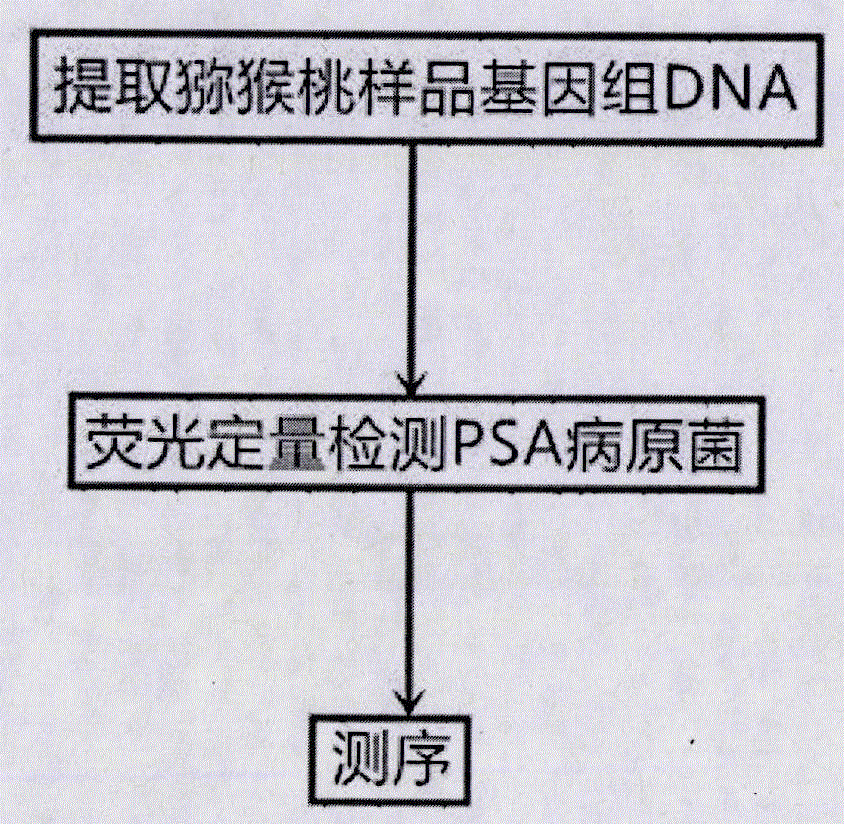
[0033] A real-time fluorescent PCR method for quantitative detection of kiwifruit canker bacterium, comprising the following steps:
[0034]Step a, extract the kiwifruit sample genomic DNA; electrophoresis detection of the band size and purity of the kiwifruit sample genomic DNA; in the step a, adopt the plant genome extraction kit to extract the kiwifruit sample genomic DNA of the suspected diseased kiwifruit branches or leaves, extract The purity of the genomic DNA band of the kiwifruit sample was detected by a micro-nucleic acid analyzer, and used for the fluorescent quantitative PCR amplification template in step b.
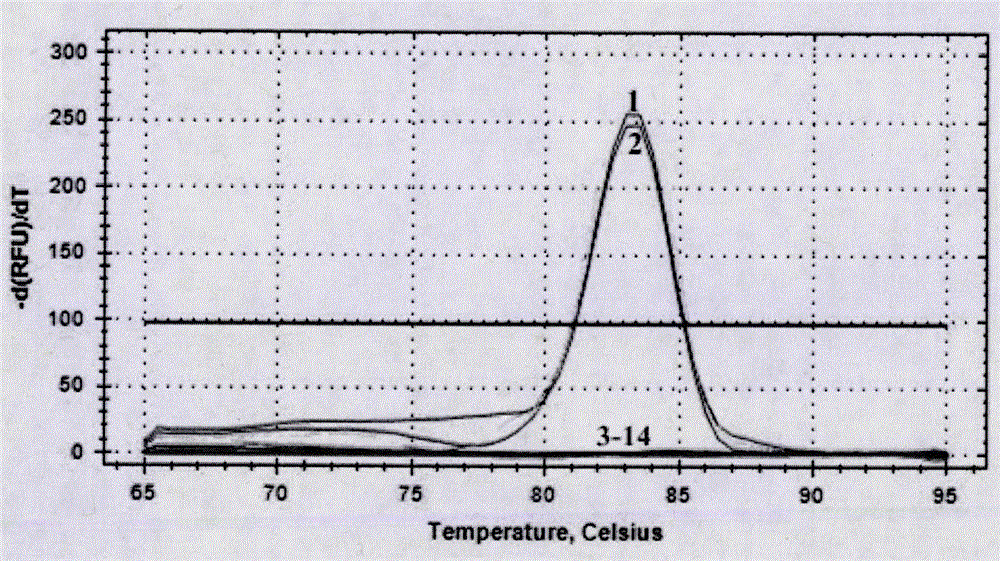
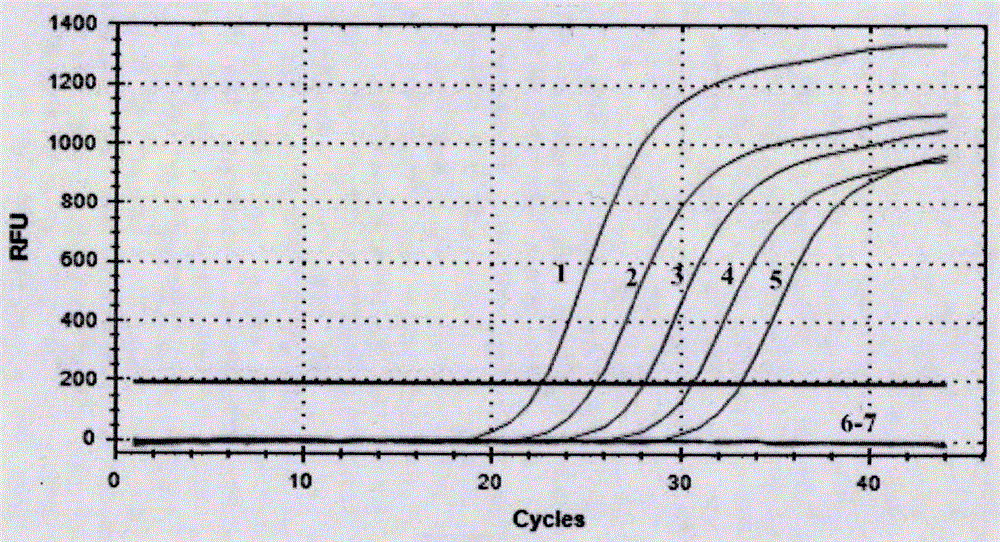
[0035] Step b, fluorescent quantitative detection of PSA pathogenic bacteria; perform fluorescent quantitative PCR amplification based on specific primers, and preliminarily determine whether PSA pathogenic bacteria are contained according to the amplification curve; the specific primers in the step b include upstream primer P3F: 5'-GGTTTCGGACACCGCAGGTTTCTACCG...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com