Promoter of 15kDa selenoprotein gene and core area and application of promoter
A promoter and gene technology, applied in the field of genetic engineering, can solve the problem of no reporter promoter region and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1、15
[0069] The cloning of embodiment 1,15kDa selenoprotein gene promoter sequence
[0070] 1. Extraction of Human Genomic DNA
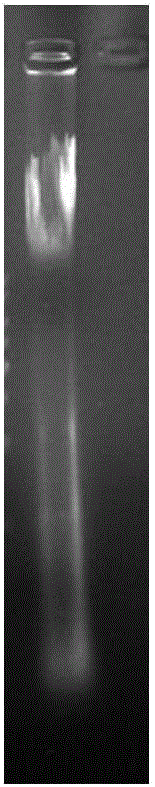
[0071] Genomic DNA was extracted from HEK293T cells by using a DNA extraction kit, the concentration of DNA detected by a nucleic acid protein detector was 0.450 μg / μl, and the value of A260 / A280 was 1.818, and 0.8% agarose gel electrophoresis was carried out as figure 1 shown. It can be seen from the figure that although a small part of the DNA is degraded, the overall quality is good and can be used for subsequent experiments.
[0072] 2. Obtaining fragments of Sep15 promoter by PCR
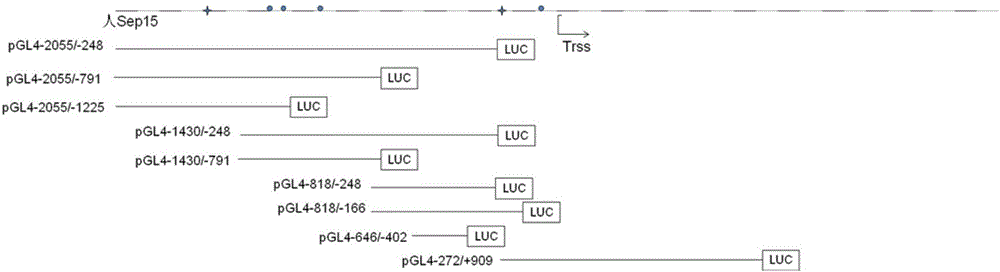
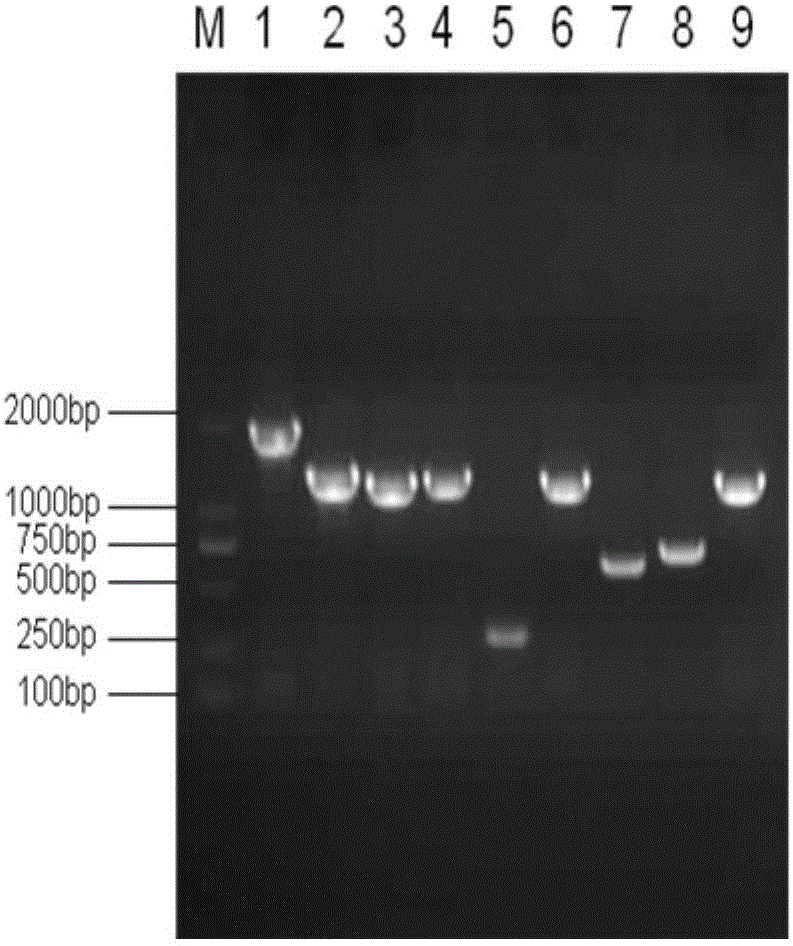
[0073] Use specific primers to clone 10 segments of the Sep15 gene promoter respectively. The names, sizes and primers of the cloned segments are shown in Table 9. The corresponding positions of the cloned segments on the promoter are shown in Table 9. figure 2 shown. Using the human genomic DNA obtained in step 1 as a template, PCR reaction was carried out, and the p...
Embodiment 2、15
[0085] Embodiment 2, the activity detection of 15kDa selenoprotein gene promoter and the determination of core region
[0086] Respectively, the recombinant plasmids of different segments of the Sep15 promoter constructed in the above-mentioned Example 1 and the pRL-TK plasmid were co-transferred into HEK293T and Neuro 2a cells at a ratio of 19:1 (such as 380ng:20ng), and pGL4 .10 [LUC2] Empty vector was used as a control, and the dual-luciferase activity was detected 24 hours after transfection, and the specific operation was carried out in the instructions of the dual-luciferase reporter gene system detection kit.
[0087] The result is as Figure 8 and Figure 9 shown. It can be seen from the figure that the enzyme activities of pGL4-2055 / -791, pGL4-2055 / -1225, pGL4-646 / -402, and pGL4-272 / +909 are close to the background value of empty pGL4.10[LUC2], that is, basically no Compared with the empty vector control, the enzyme activity of several other promoter fragment recom...
Embodiment 3
[0090] Example 3, overexpression of HSF1 eukaryotic expression vector detection of Sep15 core promoter activity
[0091] After the HEK293T cells were cultured to 80%-90% density, the cells were counted, and 0.5-2×10 5 cells were plated in a 24-well plate, and the next day, the recombinant pGL4-818 / -248 and pcDNA3.1-HSF1 were co-transfected into HEK293T cells with the pRL-TK plasmid at a ratio of 1:1 (mass ratio), and pcDNA3. 1 / Myc-His A empty vector was used as the control group. After 24 hours of transfection, the cells were lysed and the dual luciferase activity was detected.
[0092] Wherein, the specific construction process of the pcDNA3.1-HSF1 vector is as follows: using the CDS sequence of HSF1 shown in sequence 3 in the artificially synthesized sequence listing as a template, using primers HSF1-MYC-F and HSF1-MYC-R (see Table 4) PCR amplification was carried out, and the resulting PCR product was double-enzyme-digested with EcoRI and HindIII, and the enzyme-digested p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com