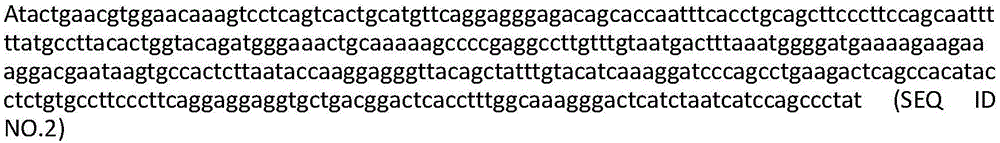TCR (T Cell Receptor) for identifying MAGE-A1 (Melanoma antigen A1) oligopeptides
A cell receptor and cell technology, applied in the field of TCR, can solve problems such as normal cell damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0145] Example 1 Cloning of MAGE-A1 Antigen Short Peptide-Specific T Cells
[0146] Peripheral blood lymphocytes (PBL) from healthy volunteers with the genotype HLA-A0201 were stimulated with the synthetic short peptide KVLEYVIKV (SEQ ID NO.:9; Beijing Saibaisheng Gene Technology Co., Ltd.). The KVLEYVIKV short peptide was annealed with biotin-labeled HLA-A0201 to prepare pHLA haploids. These haploids were combined with PE-labeled streptavidin (BD Company) to form PE-labeled tetramers, and the tetramers and anti-CD8-APC double-positive cells were sorted. Sorted cells were expanded and subjected to secondary sorting as described above, followed by monoclonalization by limiting dilution. Monoclonal cells were stained with tetramers, and the double-positive clones screened were as follows: image 3 shown.
Embodiment 2
[0147] Example 2 Construction of TCR gene and vector for obtaining MAGE-A1 antigen short peptide-specific T cell clone
[0148] with Quick-RNA TM MiniPrep (ZYMO research) extracted the total RNA of the short antigenic peptide KVLEYVIKV-specific and HLA-A0201-restricted T cell clones screened in Example 1. The cDNA was synthesized using clontech's SMART RACE cDNA amplification kit, and the primers used were designed at the C-terminal conserved region of the human TCR gene. The sequence was cloned into T vector (TAKARA) for sequencing. It should be noted that this sequence is complementary and does not contain introns. After sequencing, the sequence structures of the TCR α chain and β chain expressed by the double-positive clone are shown in Figure 1 and Figure 2, respectively. Figure 1a , Figure 1b , Figure 1c , Figure 1d , Figure 1e and Figure 1f They are the amino acid sequence of TCRα chain variable domain, the nucleotide sequence of TCRα chain variable domain,...
Embodiment 3
[0158] Example 3 Expression, refolding and purification of MAGE-A1 antigen short peptide-specific soluble TCR
[0159] In order to obtain a soluble TCR molecule, the α and β chains of the TCR molecule of the present invention may only include their variable domains and part of the constant domains respectively, and a cysteine residue is introduced into the constant domains of the α and β chains respectively To form an artificial interchain disulfide bond, the positions of the introduced cysteine residues are Thr48 of TRAC*01 exon 1 and Ser57 of TRBC2*01 exon 1; the amino acid sequence and nucleotides of the α chain sequence as Figure 4a and Figure 4b As shown, the amino acid sequence and nucleotide sequence of its β chain are as follows Figure 5a and Figure 5b The introduced cysteine residues are shown in bold and underlined letters. By the standard method described in > (Molecular Cloning a Laboratory Manual) (third edition, Sambrook and Russell), the target gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com