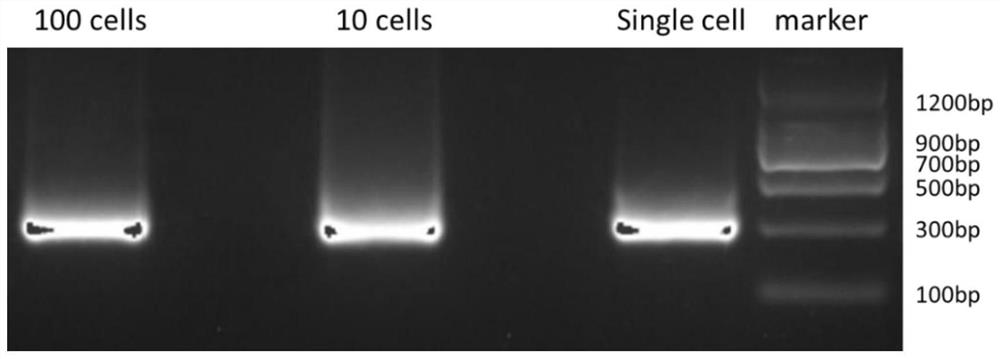
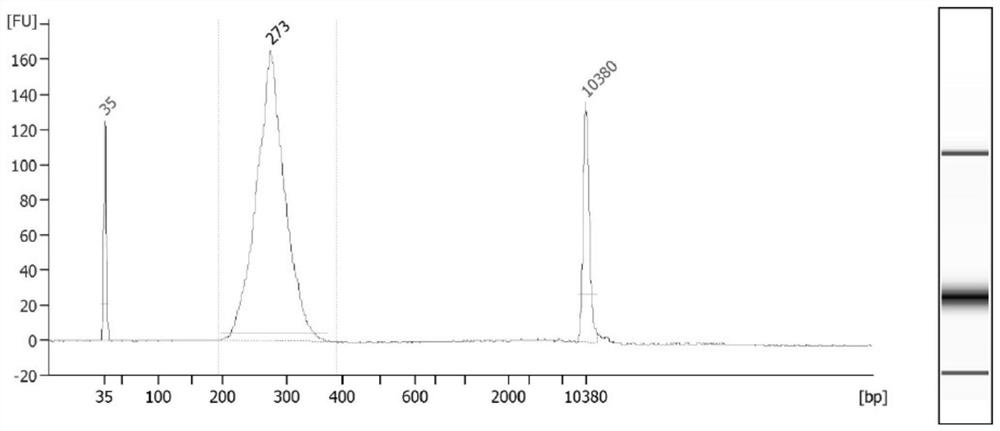
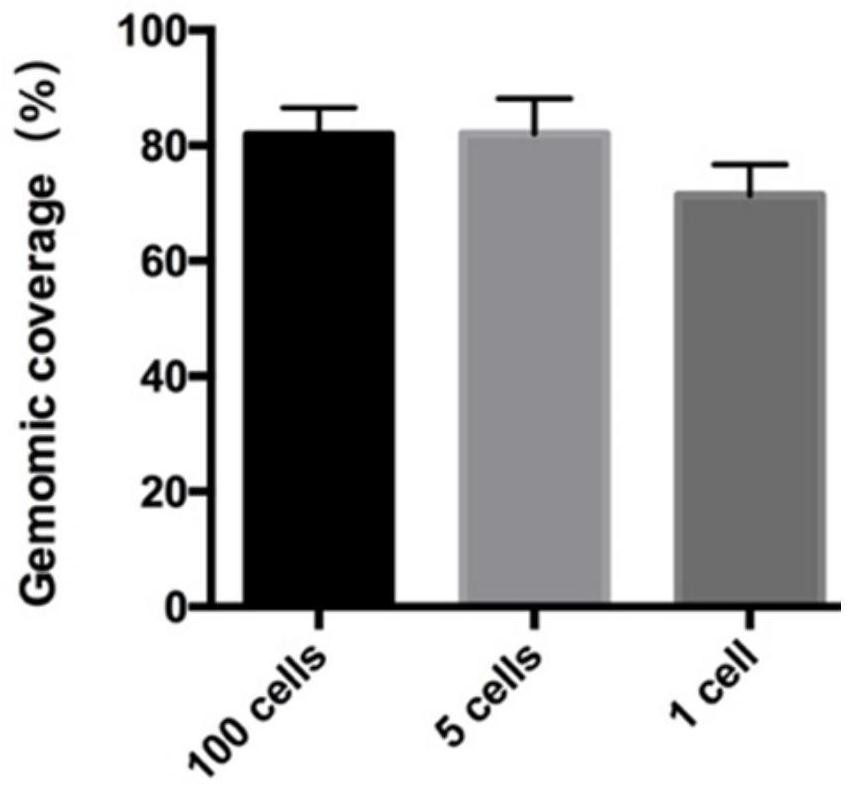
A method to detect nucleosome arrangement on the genome at the single-cell level
A genome and nucleosome technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as difficulties, heterogeneity of gene expression in cell populations, and interaction of transcription factors, so as to reduce losses, Effects of reducing DNA purification times and improving construction efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: MNase-seq detection of a small amount or a single mouse embryonic stem cell
[0031] 1. Purpose of the experiment:
[0032] The method of the invention is used to detect the nucleosome arrangement of a single or a small number of embryonic stem cells, and construct a DNA library that can be used for detection on an illumina machine.
[0033] 2. Experimental method:
[0034] 1. Cell acquisition
[0035] 1) The mouse embryonic stem cell line R1 was purchased from American Type Culture Collection (ATCC), and cultured and passaged in the laboratory.
[0036] 2) Pick a mouse embryonic stem cell clone with a mouth pipette, and wash it once in Duchenne's phosphate buffered solution (DPBS).
[0037] 3) Place the clone in trypsin at 37°C for 5 minutes.
[0038] 4) Blow and aspirate the clone repeatedly with a mouth pipette with a diameter of about 10 μm until all the cells in the clone are scattered into single cells.
[0039] 5) Wash the cells three times in pho...
Embodiment 2
[0082] Example 2: MNase-seq detection of prokaryotic
[0083] 1. Purpose of the experiment:
[0084] The method of the invention is used to detect the arrangement of nucleosomes in prokaryotes at the early stage of fertilization, and to construct a DNA library that can be used for detection on an illumina machine. In this way, it is tested whether the method of the present invention is applicable to the nucleosome arrangement detection of the haploid genome.
[0085] 2. Experimental method:
[0086] 1. Acquisition of pronuclei
[0087] 1) The SPF grade C57BL / 6 mice used in the experiment were purchased from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd., and were bred in the Experimental Animal Center of Tongji University.
[0088] 2) Take 8-10 week-old female mice, intraperitoneally inject 5U of pregnant horse serum gonadotropin (PMSG), and 48 hours later intraperitoneally inject 6U of human chorionic gonadotropin (hCG). The hormone-injected female mice we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com