System for efficient screening of candidate sgRNAs for CRISPR/CAS9 gene editing system from plants and application
A technology of CAS9 and testing system, applied in the direction of DNA/RNA fragments, genetic engineering, recombinant DNA technology, etc., can solve the problem of inability to directly reflect the real cleavage of sgRNA target sites, low cleavage efficiency of sgRNA target sites, and inability to achieve high Throughput screening and other issues, to achieve the effect of convenient construction method, low cost and high detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0023] Experimental Example 1: Screening Arabidopsis Rubisco small subunit gene candidate sgRNA
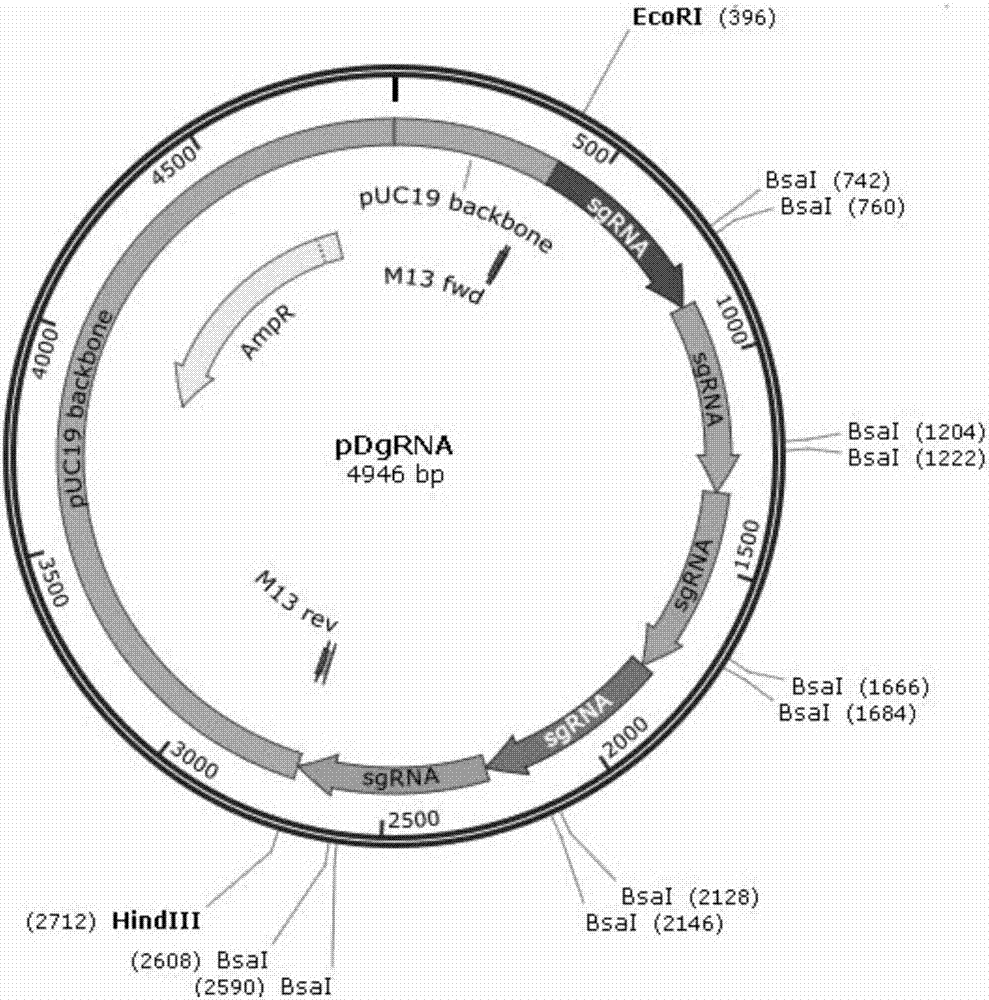
[0024] Use the CRISPR / CAS9 target site design website of Huazhong Agricultural University http: / / cbi.hzau.edu.cn / to select 5 high-scoring target sites for the Rubisco small subunit gene prediction candidate sgRNA target sites, and use edge cutting to select 5 target sites The 5 target sites were ligated into the pDgRNA vector by side-joining, and the sequence of the recombinant plasmid was confirmed by sequencing. The target sequence is as follows:
[0025] sgRNA target sequence 1: GTCGTTGTTAGCCTTGCGGGTGG
[0026] sgRNA target sequence 2: CGTGAGCACGGTAACTCACCCGG
[0027] sgRNA target sequence 3: ATAGAATATGTCTCGCAAACCGG
[0028] sgRNA target sequence 4: GGAGTCGGTGCAACCGAACAAGG
[0029] sgRNA target sequence 5: CGGAATCGGTAAGGTCAGGAAGG
[0030]Arabidopsis protoplasts were isolated using the Plant Protoplast Extraction Kit from Berelli Biotechnology Co., Ltd. The above plasmids...
experiment example 2
[0031] Experimental example 2: Screening candidate sgRNA for rice PAPST1 gene
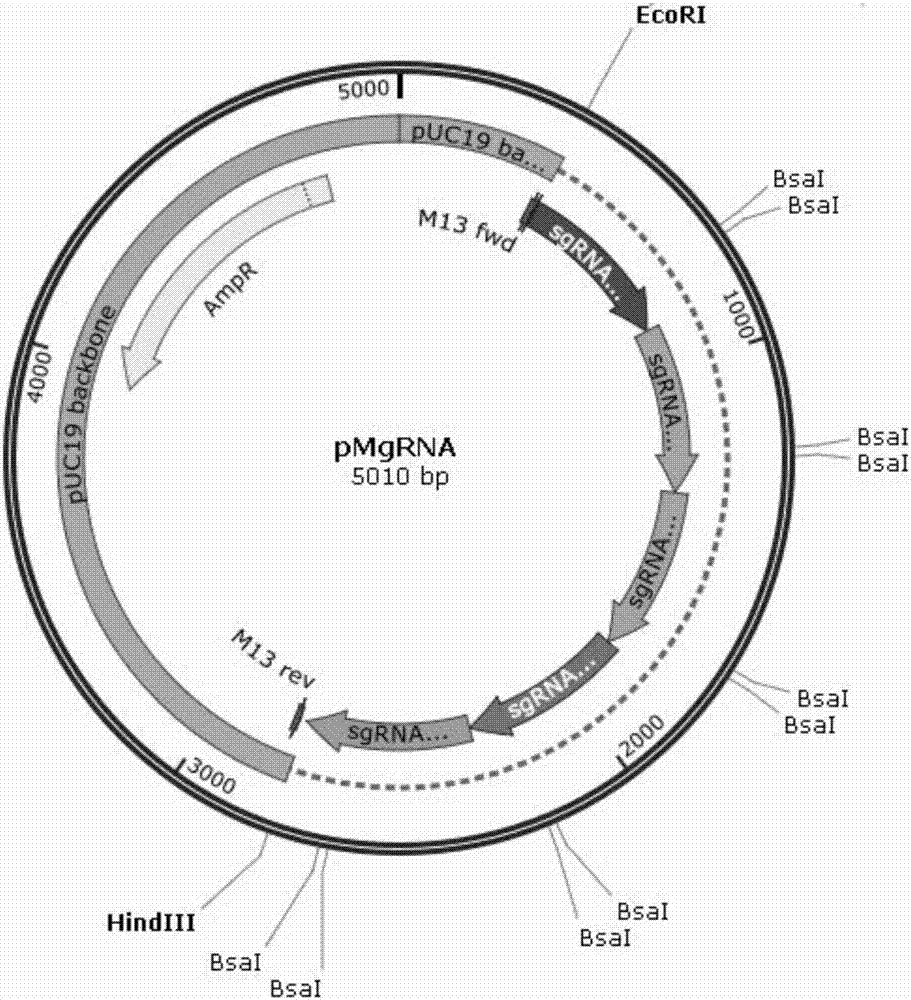
[0032] Use the CRISPR / CAS9 target site design website of Huazhong Agricultural University to select 5 high-scoring target sites of rice PAPST1 gene candidate sgRNA target sites, and connect them into the pMgRNA vector by cutting and connecting them , and the sequence of the recombinant plasmid was confirmed by sequencing. The target sequence is as follows:
[0033] sgRNA target sequence 1: CCGCATAGTTCCTTACAGTGCGG
[0034] sgRNA target sequence 2: CCGCACTGTAAGGAACTATGCGG
[0035] sgRNA target sequence 3: ACATCATCAGGTTACCTCGAGG
[0036] sgRNA target sequence 4: CATGAATCAAGTCTTCGGACTGG
[0037] sgRNA target sequence 5: GCATCCAAAACCGTGTTGTAGGG
[0038] Rice leaf sheath protoplasts were isolated using the Plant Protoplast Extraction Kit from Berelli Biotechnology Co., Ltd. The above plasmids and the CAS9 expression vector pUCCAS9 were simultaneously transformed into rice protoplasts. Genomic DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com