Constant temperature nucleic acid amplification reaction reagent
A nucleic acid amplification reaction and reagent technology, which is applied in the field of constant temperature nucleic acid amplification reaction reagents, can solve problems such as low yield of amplification products, high requirements for primers, and high purity requirements for amplified template nucleic acids, so as to improve amplification efficiency , low requirements, and high-efficiency amplification effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] The addition of spermidine and formamide in the reaction system compared with no addition
[0091] Reaction system 1 (total volume 50 μL) (containing BstDNA polymerase, spermidine, formamide): the volume percentage content of glacial acetic acid is 2%; magnesium chloride 30 mM; helicase 12 ng / μL; the volume percentage content of formamide 0.2%; spermidine 1ng / μL; betaine 1ng / μL; UvsX protein 45ng / μL; UvsY protein 45ng / μL; single-chain binding protein 700ng / μL; bovine serum albumin 1.5mg / mL; dNTPs 75mM; Tris Base 15mM; pH 7.5.
[0092] Reaction system 2 (total volume 50 μL) (containing BstDNA polymerase, without spermidine and formamide): 2% glacial acetic acid by volume; 30 mM magnesium chloride; 12 ng / μL helicase; 1 ng / μL betaine ; UvsX protein 45ng / μL; UvsY protein 45ng / μL; single-chain binding protein 700ng / μL; bovine serum albumin 1.5mg / mL; dNTPs 75mM; Tris base 15mM;
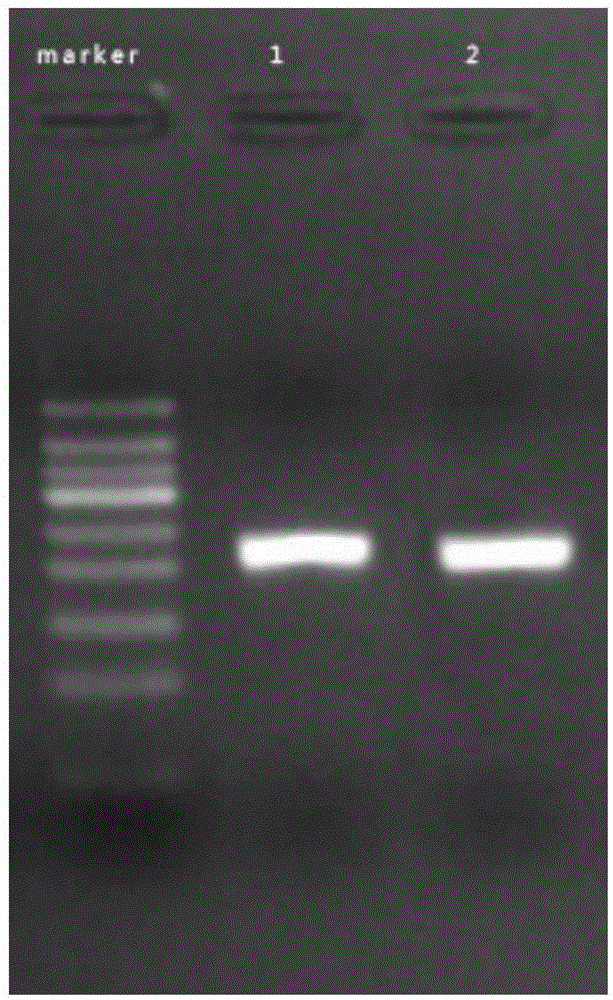
[0093] Take out 1 reagent tube 1# prefilled with 25 μL reaction system 1; and 1 reagent tube 2#...
Embodiment 2
[0096] The effect of primer design on amplification
[0097] Take out 2 reagent tubes preloaded with 25 μL reaction system one (Example 1) and mark them as 3# and 4# respectively, add 2 μL of F16-1 (20 μM) and R16-1 (20 μM) into the 3# tube, and add Add 2 μL of F16-2 (20 μM) and R16-2 (20 μM) to tube 4#; and add 5 μL of HPV16DNA template DNA I# and 1U of BstDNA polymerase to tubes 3# and 4# respectively, and then Add 13.5 μL of sterilized water to tubes 3# and 4# respectively.
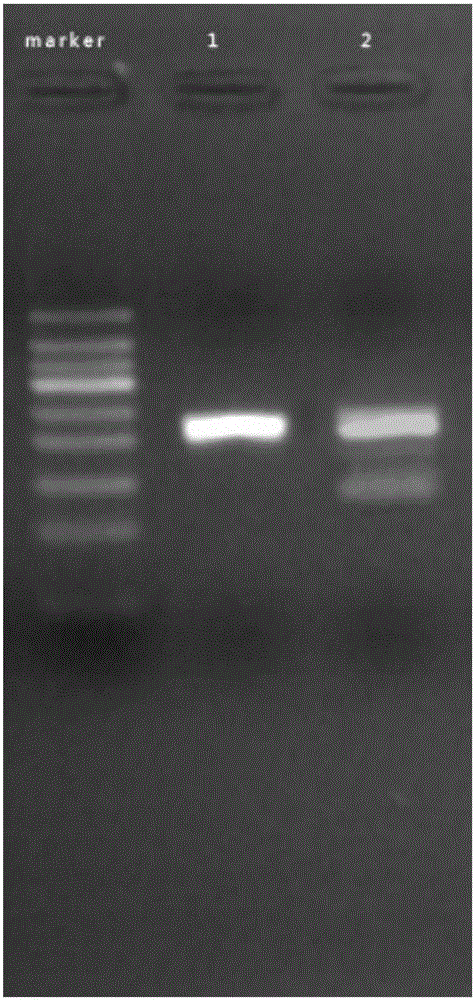
[0098]Mix tubes 3# and 4# respectively, put them into a metal bath or water bath at 37°C, and react for 15 minutes. Then take it out, add 50 μL of 1:1 phenol chloroform reagent, shake and mix well, centrifuge at 12000 for 30 seconds, take 5 μL of supernatant for agarose gel electrophoresis, it can be seen that there is one in the electrophoresis results of tubes 3# and 4# A single band of comparable brightness. see attached results figure 2 .
[0099] Take out 2 reagent tubes preloaded with 25 μL...
Embodiment 3
[0102] Requirements of Spermidine and Formamide on the Purity of Nucleic Acid Templates
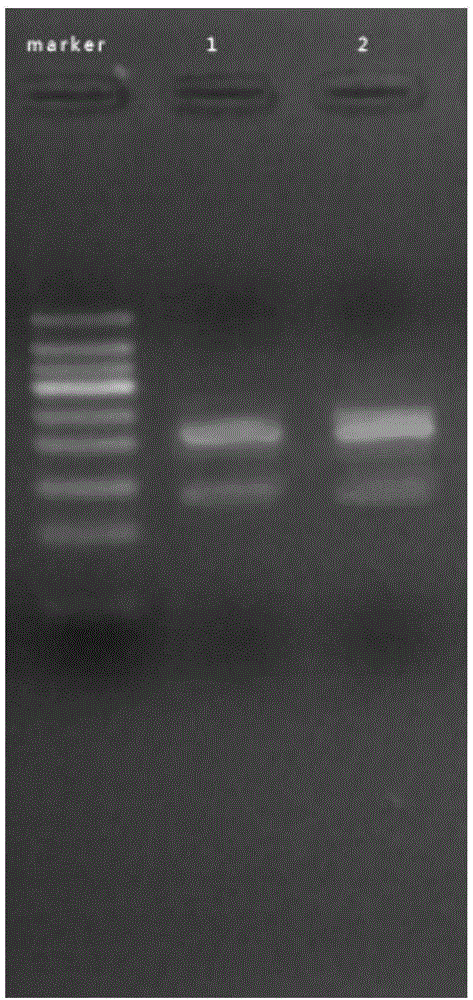
[0103] Take out 1 reagent tube 7# preloaded with 25 μL reaction system 1 (Example 1); and 1 reagent tube 8# preloaded with 25 μL reaction system 2 (Example 1), and put them into 7# and 8# tubes respectively Add 2 μL of F16-1 (20 μM) and R16-1 (20 μM); and add 5 μL of HPV16 DNA template DNA II# and 1 U of BstDNA polymerase to tubes 7# and 8# respectively, and then add sterilized water respectively 13.5μL into 7# and 8# tubes.
[0104] Mix tubes 7# and 8# respectively, put them into a metal bath or water bath at 37°C, and react for 15 minutes. Then take it out, add 50 μL of 1:1 phenol chloroform reagent, shake and mix well, centrifuge at 12000 for 30 seconds, take 5 μL of supernatant for agarose gel electrophoresis, it can be seen that there is a single band in the electrophoresis result of No. 7 tube ; while the amplification result of No. 8 tube was diffuse and no bands were seen. see ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com