Peptides having affinity for polydimethylsiloxane, and uses thereof
A technology of polydimethylsiloxane and polydimethylsiloxane materials, which is applied to peptides containing affinity tags, peptides containing His tags, peptides containing GST tags, etc., and can solve unknown problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0064] The peptide having affinity for PDMS of the present invention can be prepared by known genetic engineering technology or chemical synthesis technology. For example, a desired peptide can be produced by inserting a polynucleotide encoding the peptide shown in any one of SEQ ID NOS. 1 to 9 into a vector, and then culturing a transformant into which the vector has been introduced. The peptide having an affinity for PDMS of the present invention can also be obtained by isolating and purifying the peptide from a microorganism capable of producing the peptide. The peptide having an affinity for PDMS of the present invention can also be synthesized by known chemical synthesis techniques based on the amino acid sequence shown in any one of SEQ ID NOS. 1 to 9 or the nucleotide sequence encoding it. Chemical synthesis techniques include peptide synthesis methods such as liquid-phase or solid-phase methods. A more detailed example is the steps described in the example below. In ...
Embodiment 1
[0133] According to the following steps, the parapolydimethylsiloxane (PDMS ) affinity.
[0134] 1. Process
[0135] 1-1. Elongation factor Tu (ELN) expression vector, tryptophanase (TPA) expression vector, outer membrane protein C (OMC) expression vector build
[0136] 1) DNA Purification Kit (Promega) was used to extract the chromosomal DNA of Escherichia coli BL21(DE3) (Novagen).
[0137] 2) Use the chromosomal DNA as a template, and use KOD plus ver.2PCR kit (Toyobo Co., Ltd.) to perform PCR to amplify the ELN gene.
[0138] 3) The amplified ELN gene was mixed with the pET-22 vector (Novagen Company) in a molar ratio of 3:1, and Ligation High (Toyobo Company) equal to the mixed solution was added, and then at the Nde I position The amplified ELN gene was inserted between the dot and the Not I site for cloning.
[0139] 4) Escherichia coli HST08Premium (Takara Bio Company) was transformed with the above vector, and cultured in LB-Amp agar medium overnight.
[014...
Embodiment 2
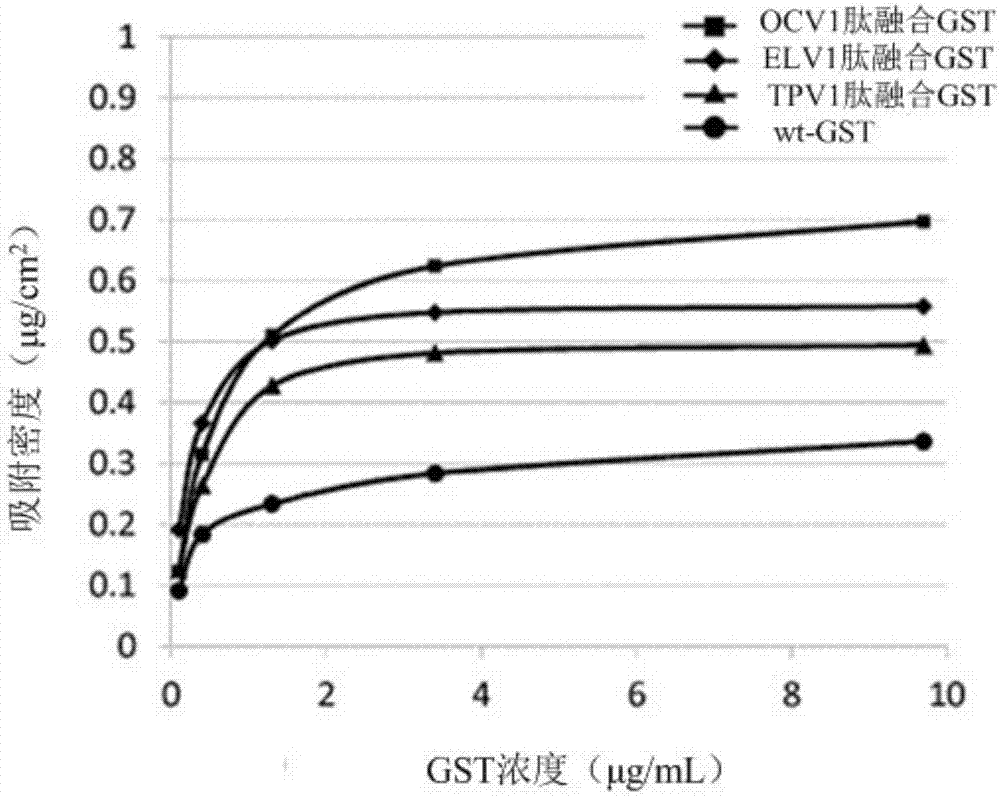
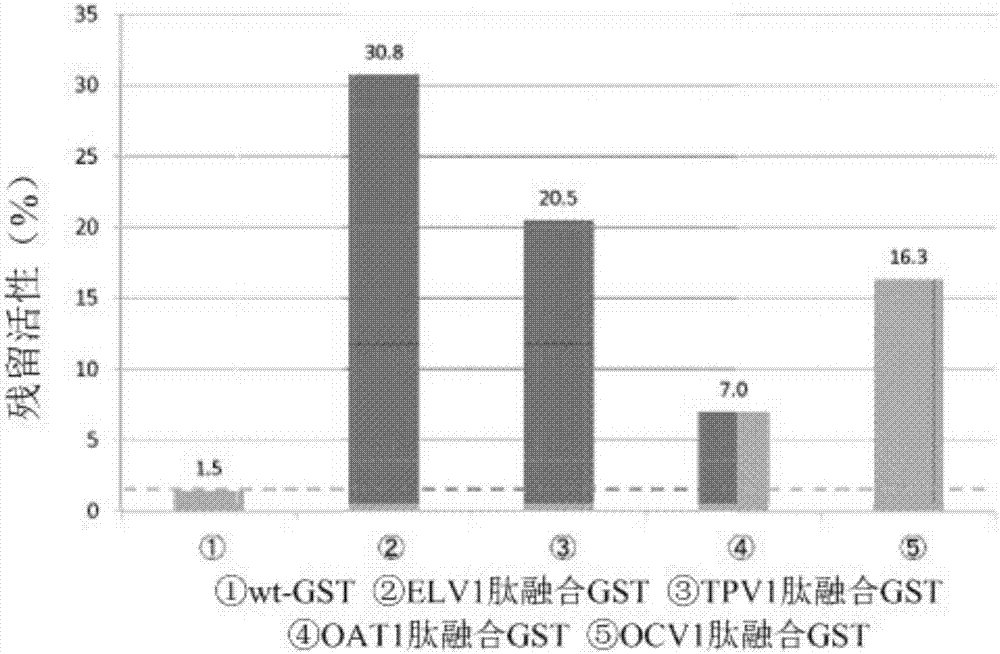
[0187] Following the steps below, studies were performed to examine whether peptide-fused GSTs immobilized to PDMS substrates maintained the original activity of GSTs.
[0188] 1. Steps
[0189] 1-1. Construction of peptide fusion GST
[0190] In this example, ELV1 peptide-fused GST, TPV1 peptide-fused GST, OCV1 peptide-fused GST, OAT1 peptide-fused GST, and wt-GST prepared in Example 1 were used.
[0191] 1-2. Determination of GST residual activity
[0192] 1. Steps
[0193] 1) Each GST solution of wt-GST, ELV1 peptide-fused GST, TPV1 peptide-fused GST, OCV1 peptide-fused GST, and OAT1 peptide-fused GST was diluted with PPB to prepare 2940 μL of each GST solution, so that after adding 30 μL of 100 mM CDNB, After 30 μL of 100 mM GSH, the GST concentration was 0.5 μg / mL.
[0194] 2) Add 30 μL of 100 mM CDNB and 30 μL of 100 mM GSH, measure the absorbance at 340 nm for 3 minutes at 25 ° C with a spectrophotometer (V-630BIO, JASCO Corporation), and calculate the chang...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com