Method for rapidly obtaining CRISPR/Cas9 (clustered regularly interspersed short palindromic repeats/Cas9) gene knockout stable cell line through monoclonal cell sorting
A gene knockout and monoclonal technology, applied in the field of genetic engineering and genetic modification, can solve the problems of low screening efficiency, poor operability, high false positive rate, etc., achieve a wide range of applications, reduce false positive rate, edit high efficiency effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] In this embodiment, the method for rapidly obtaining a CRISPR / Cas9 knockout stable cell line comprises the following steps:
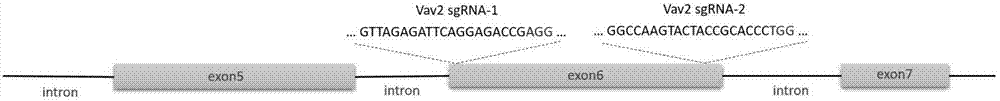
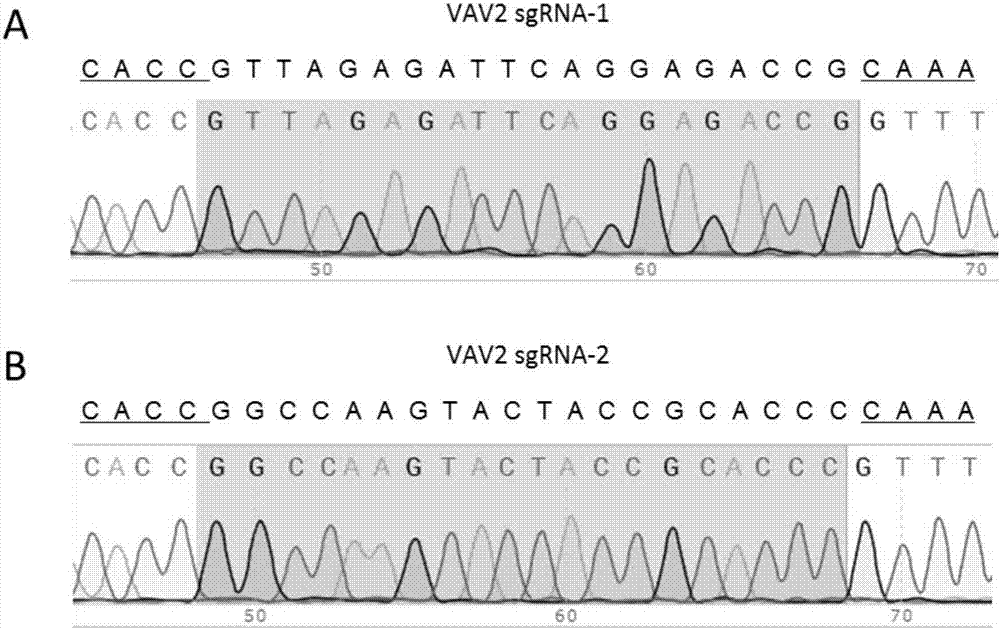
[0040] (1) Determine the specific target sites sgRNA1 and sgRNA2 of the gene Vav2 (Gene ID: 102718) to be knocked out: find the mouse Vav2 gene DNA sequence in the mouse genome database ensembl (http: / / asia.ensembl.org) (Transcript ID: ENSMUST00000056176.7), and then use the online design software CRISPOR (http: / / crispor.tefor.net / crispor.cgi) to determine the selection within the target site exon6 (exon ID: ENSMUSE00001307648) of the mouse Vav2 gene Two specific sites are used as the target sequence of sgRNA, the two target sequences are: sgRNA1 (SEQ ID NO.1): 5'-GTTAGAGATTCAGGAGACCG AGG-3', sgRNA2 (SEQ ID NO.2): 5'- GGCCAAGTACTACCGCACCC TGG-3', Vav2 gene knockout target site design as follows figure 1 shown.
[0041] (2) Design primers: According to step (1) sgRNA target sequence, design 2 pairs of 4 primers (Shanghai Bailige Biotechnology Co...
Embodiment 2
[0057] In this embodiment, the method for rapidly obtaining a CRISPR / Cas9 gene knockout stable cell line includes the following steps:
[0058](1) Determine the specific target sites sgRNA1 and sgRNA2 of the mouse gene Vav1 (Gene ID: 98923) to be knocked out: find the mouse Vav1 gene in the mouse genome database ensembl (http: / / asia.ensembl.org) DNA sequence (Transcript ID: ENSMUST00000005889), then use the online design software CRISPOR
[0059] (http: / / crispor.tefor.net / crispor.cgi), identified in the mouse Vav1 gene target site exon5 (exon ID:
[0060] ENSMUSE00000138941) select two specific sites as the target sequence of sgRNA, the two target sequences are: sgRNA1 (SEQ ID NO.9): 5'-CTACGAGGACCTAATGCGCT TGG-3', sgRNA2 (SEQ ID NO.10): 5'-CGAGGACCTTTTATGACTGCG TGG-3'.
[0061] (2) Design primers: According to step (1) sgRNA target sequence, design 2 pairs of 4 primers (Shanghai Bailige Biotechnology Co., Ltd.), and add a BbsI restriction site at the 5' end of the primer se...
Embodiment 3
[0077] In this embodiment, the method for rapidly obtaining a CRISPR / Cas9 gene knockout stable cell line includes the following steps:
[0078] (1) Determine the specific target sites sgRNA1 and sgRNA2 of the mouse gene DSG4 (Gene ID: 2661061) to be knocked out: find the mouse DSG4 gene in the mouse genome database ensembl (http: / / asia.ensembl.org) DNA sequence (Transcript ID: ENSMUST00000019426.4), and then use the online design software CRISPOR (http: / / crispor.tefor.net / crispor.cgi) to determine the target site exon12 (exonID: ENST00000308128.8) in the mouse DSG4 gene ) to select two specific sites as the target sequence of sgRNA, the two target sequences are: sgRNA1 (SEQ ID NO.17): 5'-CTTAGCCGTAAGGATTGCCG AGG-3', sgRNA2 (SEQ ID NO.18): 5'-GTGGTTGTCATCGCAATCAC AGG-3'.
[0079] (2) Design primers: According to step (1) sgRNA target sequence, design 2 pairs of 4 primers (Shanghai Bailige Biotechnology Co., Ltd.), and add a BbsI restriction site at the 5' end of the primer seq...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com