A method for high-accuracy site-specific gene knockout in Coprinus cinerea
A high-accuracy, gene-knockout technology is applied in the field of genetic engineering to reduce labor force, facilitate procurement, and improve work efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
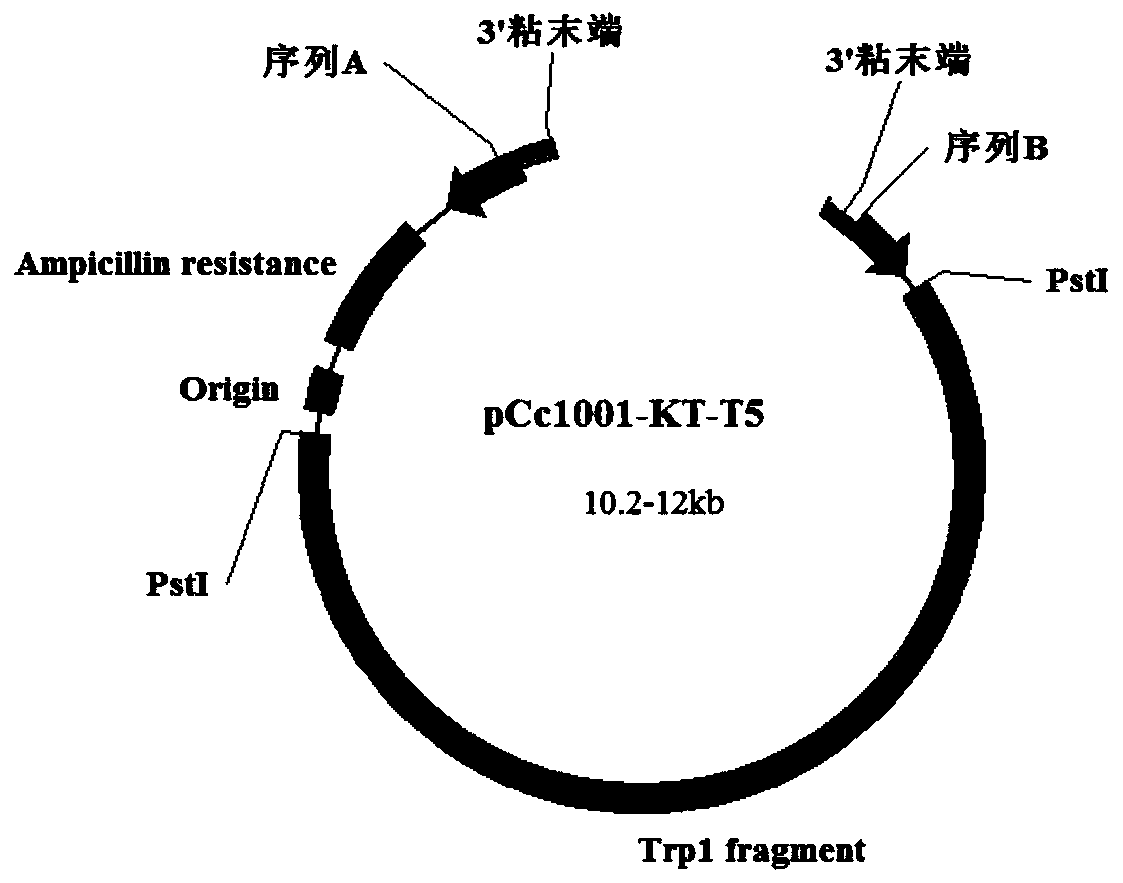
[0025] Step 1: 1000 bp homologous sequence cloning of the upstream (sequence A1) and downstream (sequence B1) of the endocellulase gene.
[0026]The polymerase chain reaction (PCR) method was used for cloning, the DNA polymerase used was high-fidelity DNA polymerase (Phusion DNA Polymerase) produced by NEB, the amplification template was Coprinus cinerea LT2 genomic DNA, and the primers were based on Coprinus cinerea. Oligonucleotides designed and synthesized from the genome sequence of okayama7#130, PPF1, PPR1, PTF1 and PTR1, the sequence information is as follows:
[0027] PPF1:5'-CTCGGTCTATTCTTTTGATTTAGATGGCACCGTTCTTGTCGGCTG-3'
[0028] PPR1:5'-GAAGATGCGGCCGCTGTTAC AGCTGGTCTACAATGTTCGAT-3'
[0029] PTF1: 5'-GTAACAGCGGCCGCATCTTC AAGGGTGAGTTCGGTTCTAT-3'
[0030] PTR1:5'-GCCGAAATCGGCAAAATCCCTTAGCTGTGGTAGGCCCCTAATCAGG-3'
[0031] Primers PPF1 and PPR1 are used to clone the 1kb region upstream of the endocellulase, and PTF1 and PTR1 are used to clone the 1kb region downstream...
Embodiment 2
[0055] Step 1: Cloning of 100 bp homologous sequences upstream (sequence A2) and downstream (sequence B2) of the endocellulase gene.
[0056] The cloning method is the same as in Example 1, the primers are PPF2, PPR1, PTF1 and PTR2, and the sequence information is as follows:
[0057] PPF1:5'-CTCGGTCTATTCTTTTGATTTAGATGGCACCGTTCTTGTCGGCTG-3'
[0058] PPR2:5'-GAAGATGCGGCCGCTGTTAC TGGGCCTGGTCATTGGGCA-3'
[0059] PTF2: 5'-GTAACAGCGGCCGCATCTTC GTTATCTTCAAGCGCCTA-3'
[0060] PTR1:5'-GCCGAAATCGGCAAAATCCCTTAGCTGTGGTAGGCCCCTAATCAGG-3'
[0061] Primers PPF1 and PPR2 are used to clone the 100bp region upstream of the endocellulase, PTF2 and PTR1 are used to clone the 100bp region downstream of the endocellulase, the PCR reaction system is except that the primer PPR2 replaces PPR1, PPF2 replaces PPF1, the amount of each corresponding component With embodiment 1. The PCR amplification conditions were the same as in Example 1 except that the extension time was changed to 20 sec, and the...
Embodiment 3
[0075] Step 1: Cloning of 500 bp homologous sequences upstream (sequence A3) and downstream (sequence B3) of the endocellulase gene.
[0076] The cloning method is the same as in Example 1, the primers are PPF3, PPR1, PTF1 and PTR3, and the sequence information is as follows:
[0077] PPF1:5'-CTCGGTCTATTCTTTTGATTTAGATGGCACCGTTCTTGTCGGCTG-3'
[0078] PPR3: 5'-GAAGATGCGGCCGCTGTTAC GTAAAGACGCCTGGTGGAAGTGTCTG-3'
[0079] PTF3: 5'-GTAACAGCGGCCGCATCTTC TGCACTTATGCGCGAGCCTGGGTC-3'
[0080] PTR1:5'-GCCGAAATCGGCAAAATCCCTTAGCTGTGGTAGGCCCCTAATCAGG-3'
[0081] Primers PPF1 and PPR3 are used to clone the 500bp region upstream of the endocellulase, and PTF3 and PTR1 are used to clone the 500bp region downstream of the endocellulase. The PCR reaction system replaces PPR1 with primer PPR3 and replaces PPF1 with PPF3. With embodiment 1. The PCR amplification conditions were the same as in Example 1 except that the extension time was changed to 30 sec, and the PCR product purification metho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com