Highly catalytically active pet hydrolase mutants
A mutant, hydrolase technology, applied in hydrolase, biochemical equipment and methods, fermentation and other directions, can solve the problems of low efficiency of PET plastic degradation, insufficient catalytic function of key enzyme Ec_PETase, difficult industrial application, etc. Degradation cost, high enzymatic activity, effect of simplifying degradation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1, Escherichia coli secreted often expressed PET hydrolase (Ec_PETase) mutation hotspot screening
[0035] 1. Construction of 3D model of PETase
[0036]Enter the amino acid sequence of PETase: (GenBank accession number, GAP38373.1) into the three websites I-TASSER, MULTICOM, and ROBETTA to obtain the 3D model of PETase, use Rampage to evaluate the obtained model, and select 3 models for follow-up Docking experiment.
[0037] 2. Use Auto Dock software for substrate docking
[0038] Since the pNPB method is a commonly used lipase activity detection method, pNPB, the product of lipase hydrolysis, has an absorption peak at 405nm under the conditions of 34°C and pH=7.4. Therefore, we used pNPB as the substrate for molecular docking. GaussView was used to draw its 3D structure, and dynamic optimization was performed to finally obtain the most stable conformation of pNPB in space. The best results were obtained after docking simulation analysis.
[0039] 3 mutati...
Embodiment 2
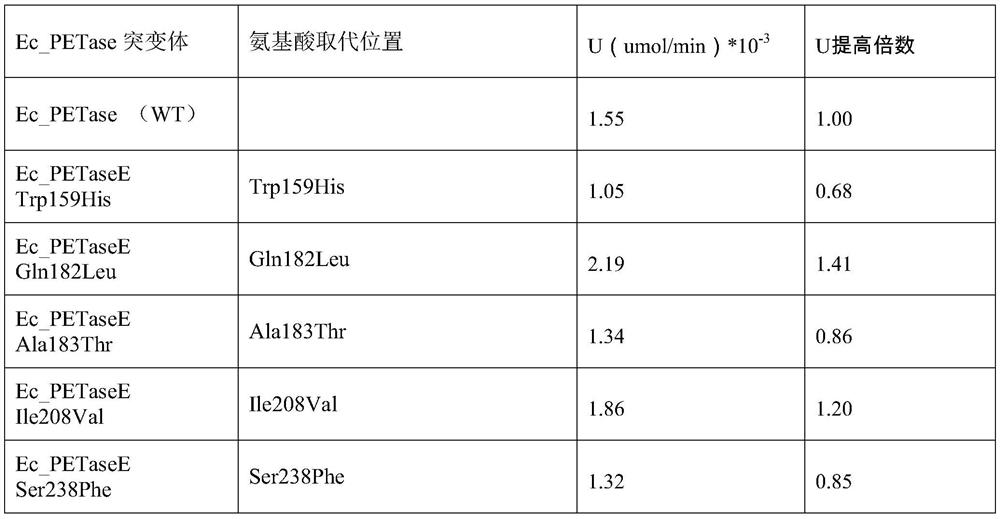
[0041] Example 2. Construction of site-directed mutants of mutation hotspots Trp159, Gln182, Ala183, Ile208 and Ser238.
[0042] The primers used in the experiment are listed in Table 1 below:
[0043] Table 1 Primers used for construction of site-directed mutants
[0044] Primer serial number Primer sequence (5'-3') W159H-Forward SEQ ID No.9 TATGGGTCATAGCATGGGTGGCGGTGGCAGC W159H-Reverse SEQ ID No.10 CACCCATGCTATGACCCATAACGCCCATACG Q182L-F SEQ ID No.11 GGCGCCGCTGGCGCCGTGGGACAGCAGCTTC Q182L-R SEQ ID No.12 CACGGCGCCAGCGGCGCCGCAGCTTTCAGGCT A183T-F SEQ ID No.13 CGCCGCAAACCCCCGTGGGACAGCAGCTTCAGC A183T-R SEQ ID No.14 TCCCACGGGGTTTGCGGCGCCGCAGCTTTCAG I208V-F SEQ ID No.15 ACGATAGCGTTGCGCCGGTGAACAGCAGCGCG I208V-R SEQ ID No.16 ACCGGCGCAACGCTATCGTTCTCGCACGCAAA S238F-F SEQ ID No.17 GCAGCCACTTCTGCGCGAACAGCGGTAACAGC S238F-R SEQ ID No. 18 GTTCGCGCAGAAGTGGCTGCCACCGTTAATTTC
[0045] The n...
Embodiment 3
[0047] Example 3, EcPETase recombinant expression and activity evaluation
[0048] 1. Take pUC57-EcPETase genetically engineered Escherichia coli mutants respectively, culture them with shaking in LB liquid medium at 37°C and 180rpm for 4 hours (OD600=1.5), centrifuge to separate the protein secreted into the extracellular space and concentrate it to 0.025mg / ml is used for the determination of enzyme catalytic activity.
[0049] 2. Obtain a concentrated supernatant sample of the overnight culture product of the pUC57-EcPETase genetically engineered E. coli strain, using pNPB as the substrate. References: "oshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K , Miyamoto K, Kimura Y, Oda K.2016.A bacterium that degrades and assimilates poly(ethylene terephthalate).Science,351(6278):1196-1199."The experimental method disclosed in the test pUC57-EcPETase genetically engineered Escherichia coli strain Catalytic activity of EcPETase in culture medium. Add 100 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com