Cloud computing acceleration method for gene sequence alignment
A gene sequence and cloud computing technology, applied in computing, sequence analysis, special data processing applications, etc., can solve the problem of gene sequence data taking a long time, and achieve the effect of easy development and maintenance, code maintenance, and good flexibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] The present invention will be further described below in conjunction with specific examples.
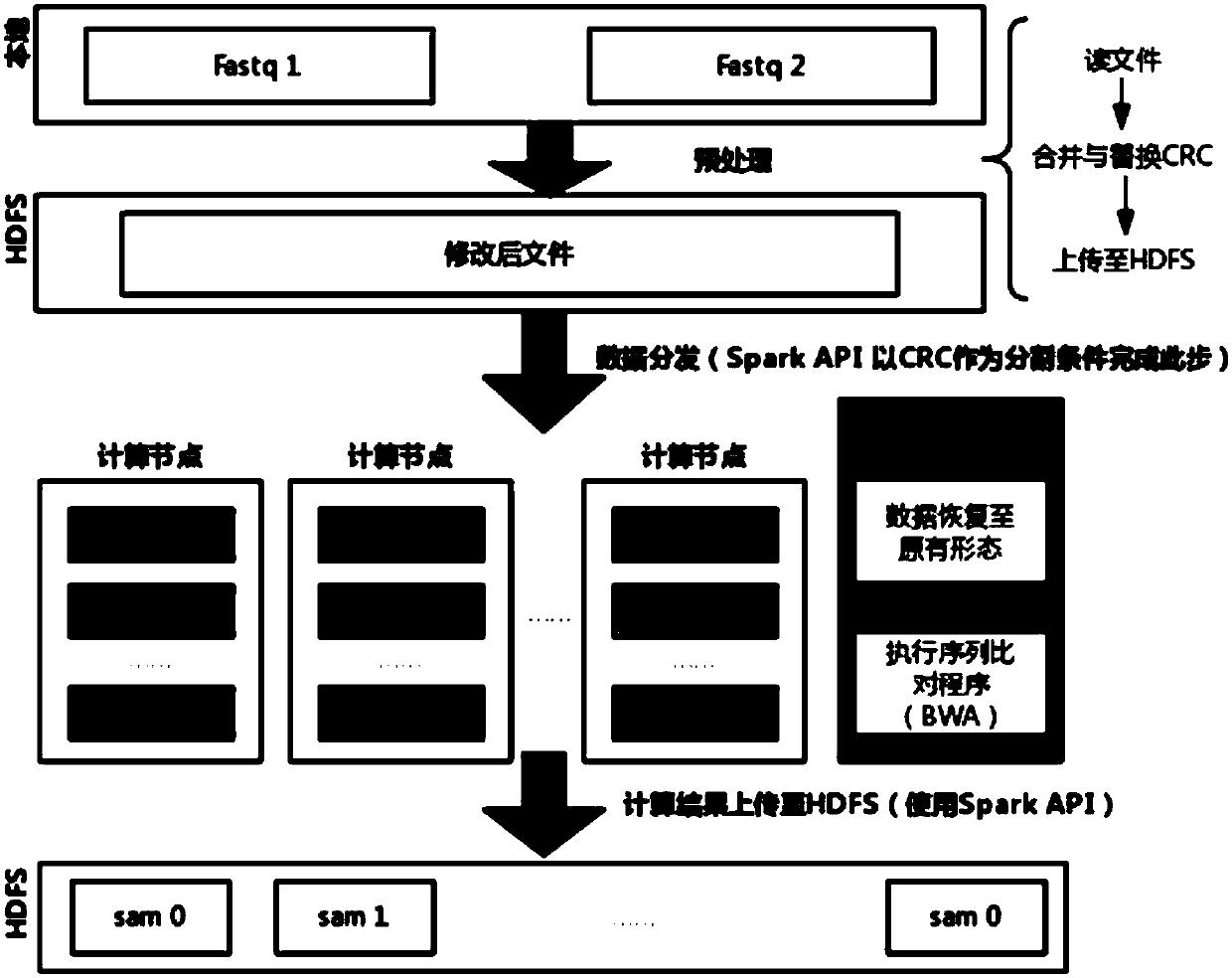
[0034] Such as figure 1 As shown, the cloud computing acceleration method for gene sequence comparison provided in this embodiment includes the following steps:
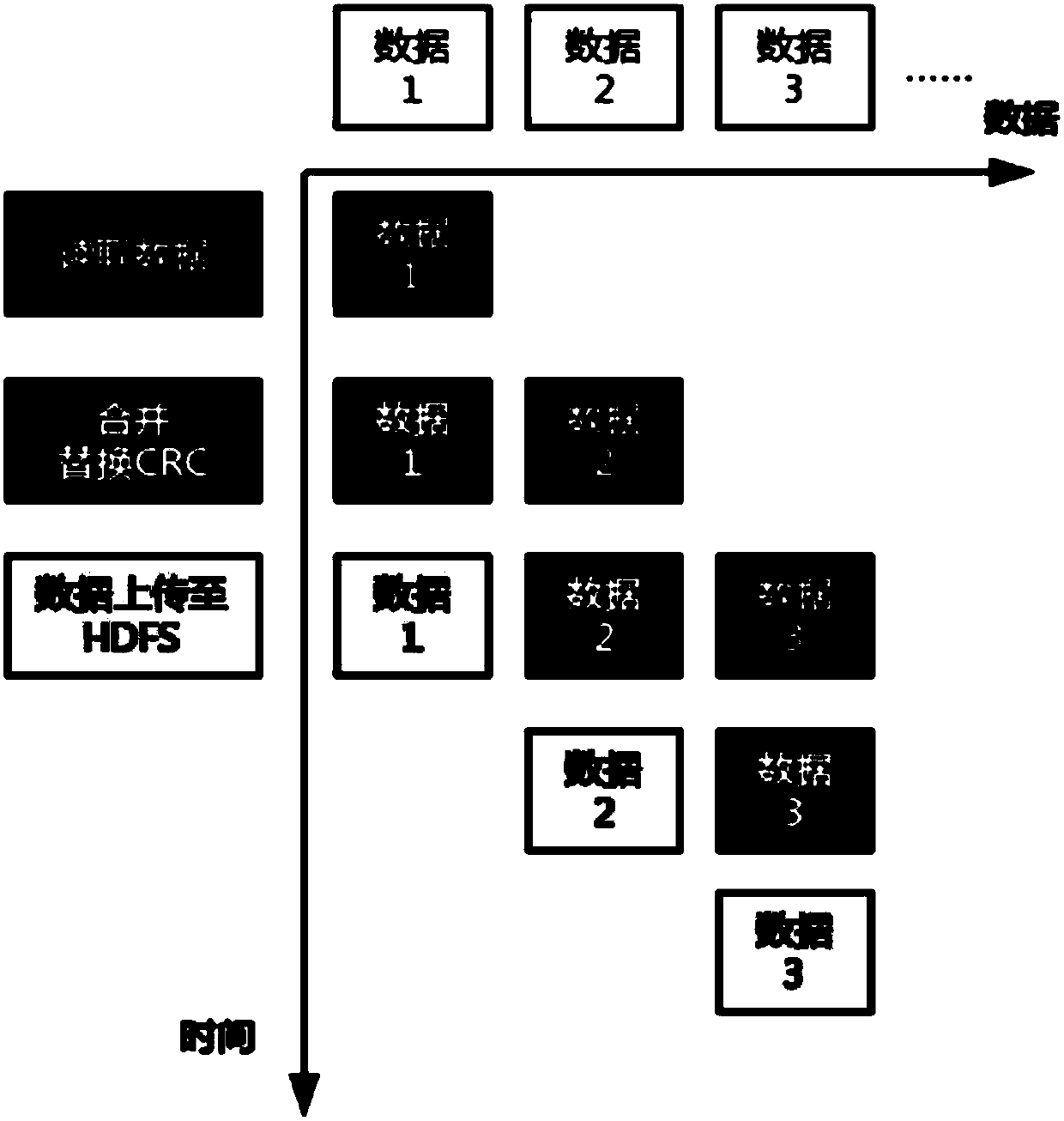
[0035] S1. Preprocess the off-machine data file Fastq of the gene sequencer to ensure the integrity of the data when the data is distributed, including reading the data, merging multiple input files and saving the data to the file system.
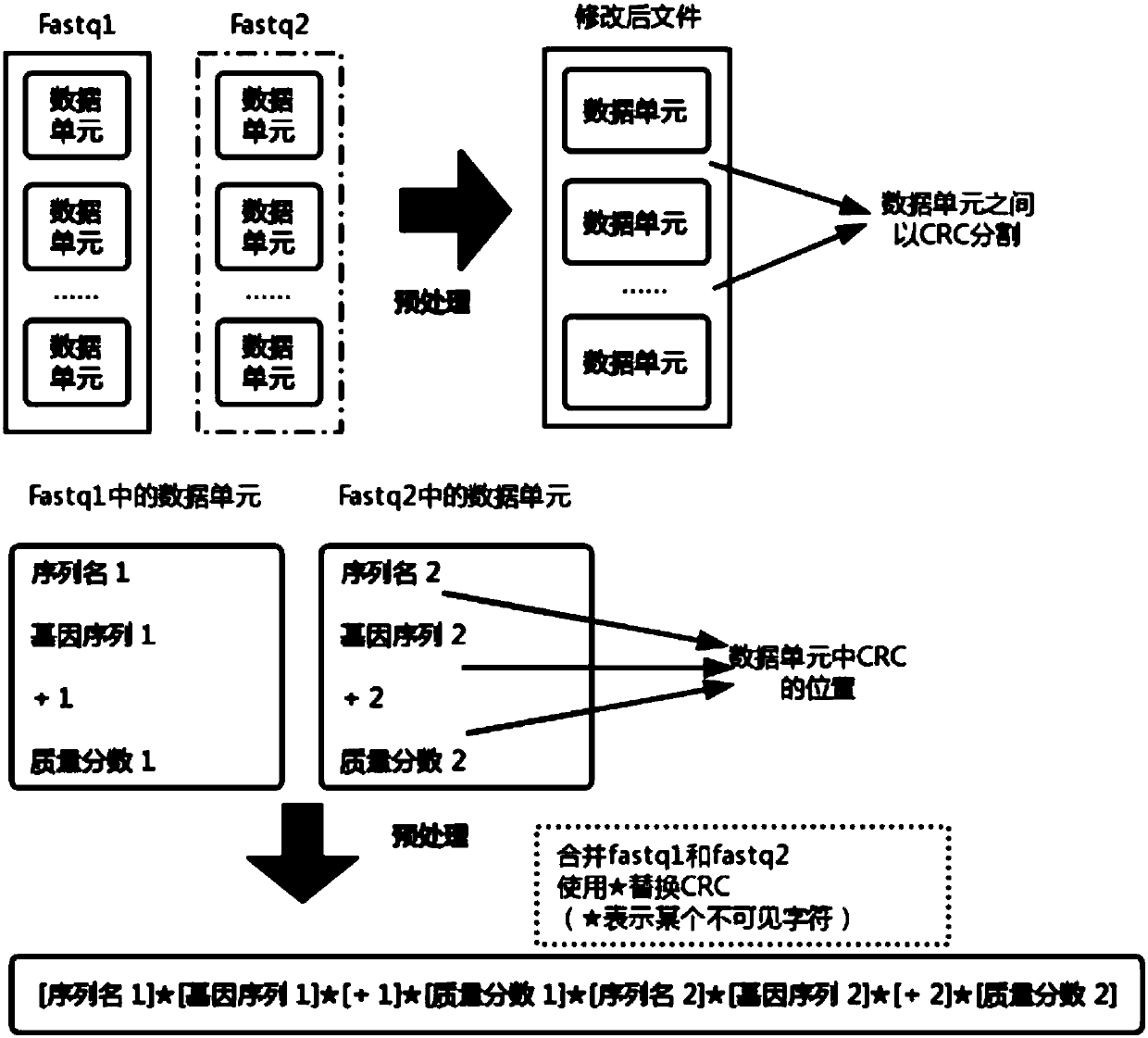
[0036] figure 2 The style of the Fastq format file and the modified file form are given in . In the Fastq file, every four lines form the complete information of a reading sequence, namely figure 2 A data unit within a Fastq file in . Paired-end sequencing produces two Fastq files, namely figure 2 Two files Fastq1 and Fastq2 in. The data units in the two Fastq files are in one-to-one correspondence, and together constitute a complete piece of information that the gene seq...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com